Being picky at the buffet (as a Verrucomicrobiota cell)
Published in Microbiology

Since I started examining metagenomic samples during my Ph.D., I always noticed that in most terrestrial and aquatic environments, Verrucomicrobiota (aka Verrucomicrobia) were always there. I remembered that this group was never the most abundant nor, at the moment, the most exciting compared to nitrifiers, which at the time were driving my curiosity(1). It was not until a few years after finishing my Ph.D. and some changes (e.g., a move from soil to marine environments) that Verrucomicrobiota was on my radar again.
Over the past few years, our group at the Max Planck Institute for Marine Microbiology has examined the annual and recurrent spring blooms in the North Sea(2, 3). These bloom events are characterized by a massive release of polysaccharides following the demise of the microalgae. This excess of ‘food’ can quickly initiate a bloom of heterotrophic bacteria that specialize in consuming these freshly available myriads of polysaccharides. Our research group has detected and identified major bacterial(2) and even archaeal (4) groups participating in the remineralization of the organic matter. Bacteroidetes, Gammaproteobacteria, and Alphaproteobacteria are among the main groups that come back every year for the 'delicious treats' that the aftermath of the algae bloom has to offer. The use of culture-independent techniques such as metagenomic sequencing has allowed us to inspect the genetic basis for the mechanisms found in these bloom-associated heterotrophic bacteria.
It was when examining these groups that Verrucomicrobiota caught my attention again. Throughout the spring blooms, this group reaches up to 8% of the total community. Another striking fact is that, compared to most bacterial clades, Verrucomicrobiota are strictly seasonal, and only specific species are present during the spring and summer blooms(5). For the rest of the years, these verrucomicrobial ‘bloom’ populations don’t reach abundances high enough to be detected by our sequencing strategies. ‘This is cool’- I thought.
Here, we found that Verrucomicrobiota cells are highly specialized consumers of ‘hard to degrade' sugars (here, I’m referring to polysaccharides containing sulfate and fucose). Suppose you were to compare the metabolic potential lightly: Verrucomicrobiota populations and prolific heterotrophs, such as Bacteroidetes, have equivalent numbers of sulfates and glycoside hydrolyse genes for polysaccharide degradation. However, it’s more than just numbers: Most Verrucomicrobiota cells carry specialized degradation pathways, that when linked to bacterial microcompartments, can circumvent the accumulation of the toxic lactaldehyde. No other group in the bloom unites such specialized mechanisms for the degradation of fucose and rhamnose and can reach relative abundances over 8% of the total community.
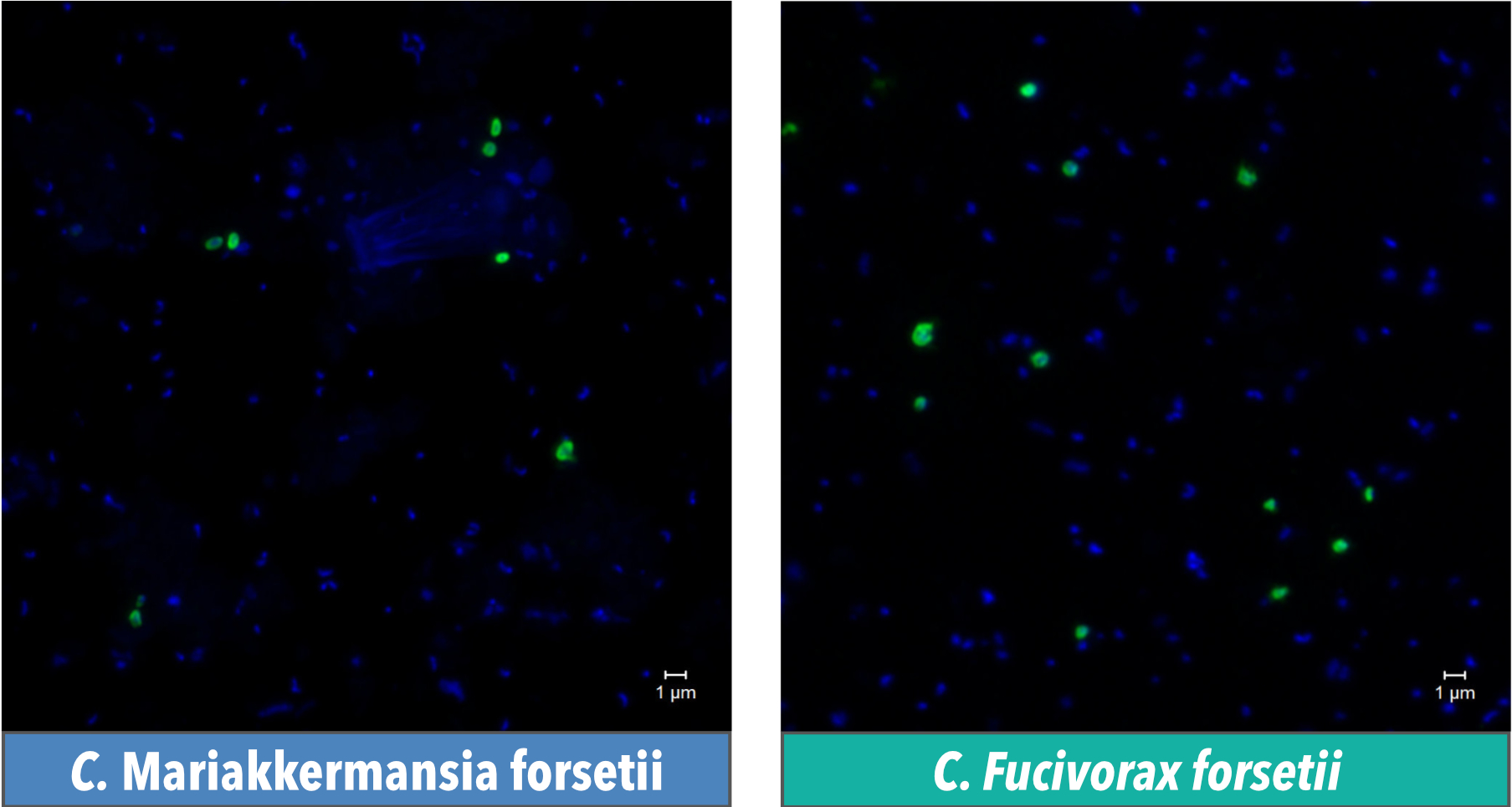
After looking at too many text files for hours, it is a big relief knowing that what we predicted as metagenomic-assembled genomes (MAGs) can actually being seen under the microscope using CARD-FISH. Modified from L.H. Orellana at al., 2021 ISMEJ Creative Commons - CC BY 4.0
We also named here two Verrucomicrobiota members: Candidatus Mariakkermansia forsetii and Candidatus Fucivorax forsetii - we can see them!
In summary, it seems that being picky about what they eat (but being great at it) makes this group successful and recurrent every year during the spring blooms.
See the paper here: Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms
References
1. Orellana LH, Chee-Sanford JC, Sanford RA, Löffler FE, Konstantinidis KT. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl Environ Microbiol. 2017;84(2):1646-17.
2. Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336(6081):608-11.
3. Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L, et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. Elife. 2016;5:e11888.
4. Orellana LH, Ben Francis T, Kruger K, Teeling H, Muller MC, Fuchs BM, et al. Niche differentiation among annually recurrent coastal Marine Group II Euryarchaeota. ISME J. 2019;13(12):3024-36.
5. Chafee M, Fernàndez-Guerra A, Buttigieg PL, Gerdts G, Eren AM, Teeling H, et al. Recurrent patterns of microdiversity in a temperate coastal marine environment. ISME J. 2018;12(1):237-52.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in