Cracking the Genetic Code of Glioblastoma: The Hidden Role of HOX Genes
Published in Neuroscience and Genetics & Genomics
🔬 Why HOX Genes?
HOX (Homeobox) genes are essential during embryonic development. Like master architects, they determine where the head, tail, arms, and legs form. These genes are highly conserved across species and play a crucial role in organizing the body’s structure along the anterior-posterior axis.
But in recent years, researchers have discovered that HOX genes can be reactivated or dysregulated in cancer—and not just any cancer. In glioblastoma (GBM), one of the deadliest forms of brain tumor, abnormal HOX gene expression has been linked to:
-
Tumor growth
-
Therapeutic resistance
-
Poor survival outcomes
This unexpected connection between developmental biology and cancer progression caught our attention.
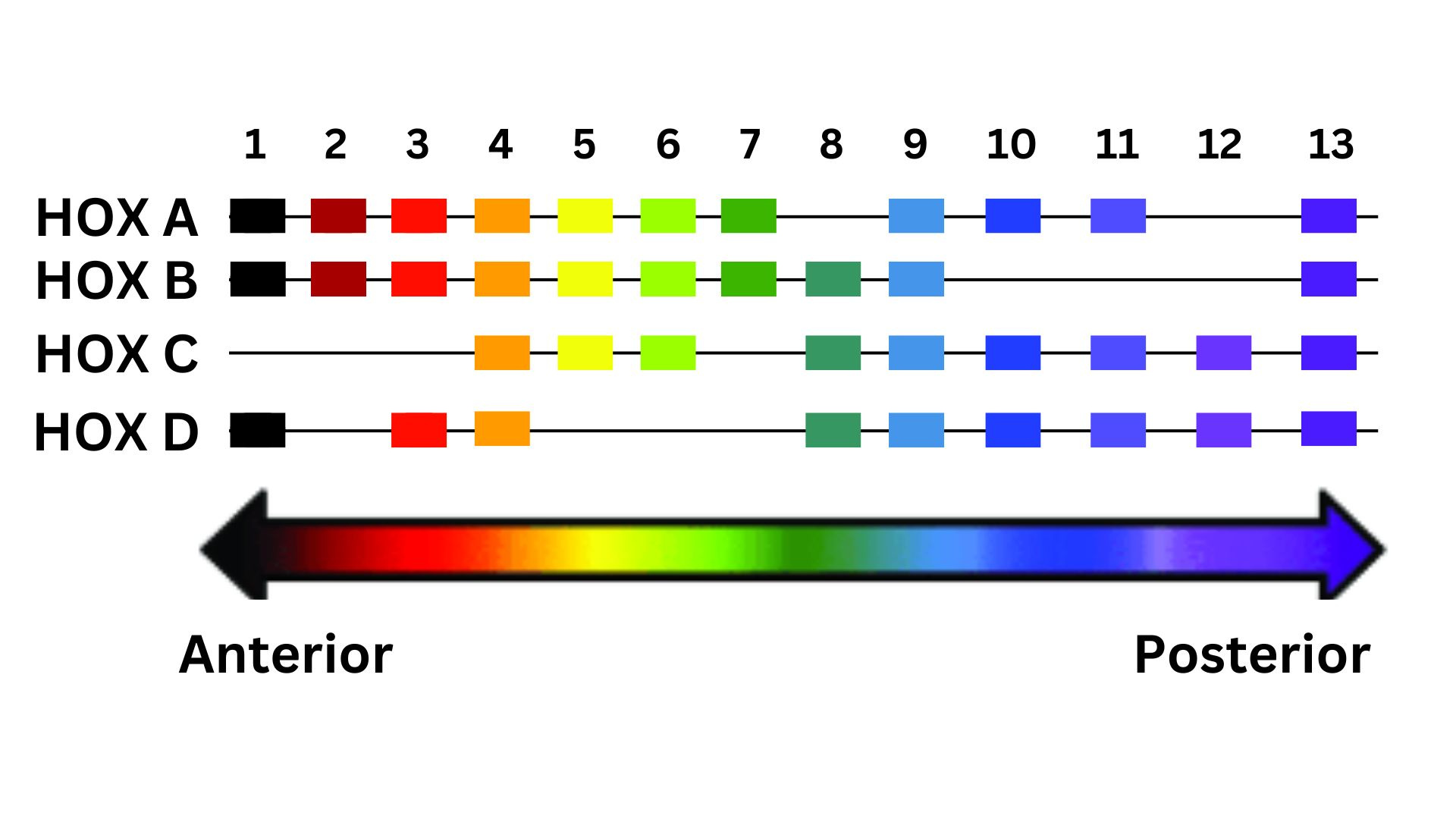
The genomic architecture of the HOX family is present in each of the four loci. Humans have four loci of HOX genes: HOXA contains 11 genes, located in chromosome 7p15.2; HOXB contains 10 genes, located in chromosome 17q21.3; HOXC contains 9 genes, located in chromosome 12q13.3; HOXD contains 9 genes, located in the chromosome 2q31 (p represent the short arm of the chromosome, q represent the long arm of the chromosome). During embryonic development, HOX genes are expressed sequentially in partially overlapping zones along the anterior-posterior axis.
🧠 The Glioblastoma Challenge
Glioblastoma is an aggressive, fast-growing tumor with a notoriously poor prognosis. Despite surgery, radiation, and chemotherapy, the average survival time remains just 12–15 months after diagnosis.
We wondered—could HOX genes be one of the hidden drivers behind glioblastoma’s resistance and rapid progression?
🧾 What We Did?
We conducted an in-depth review of the scientific literature to answer this question. Our article:
-
Summarizes the current knowledge about HOX gene expression in GBM
-
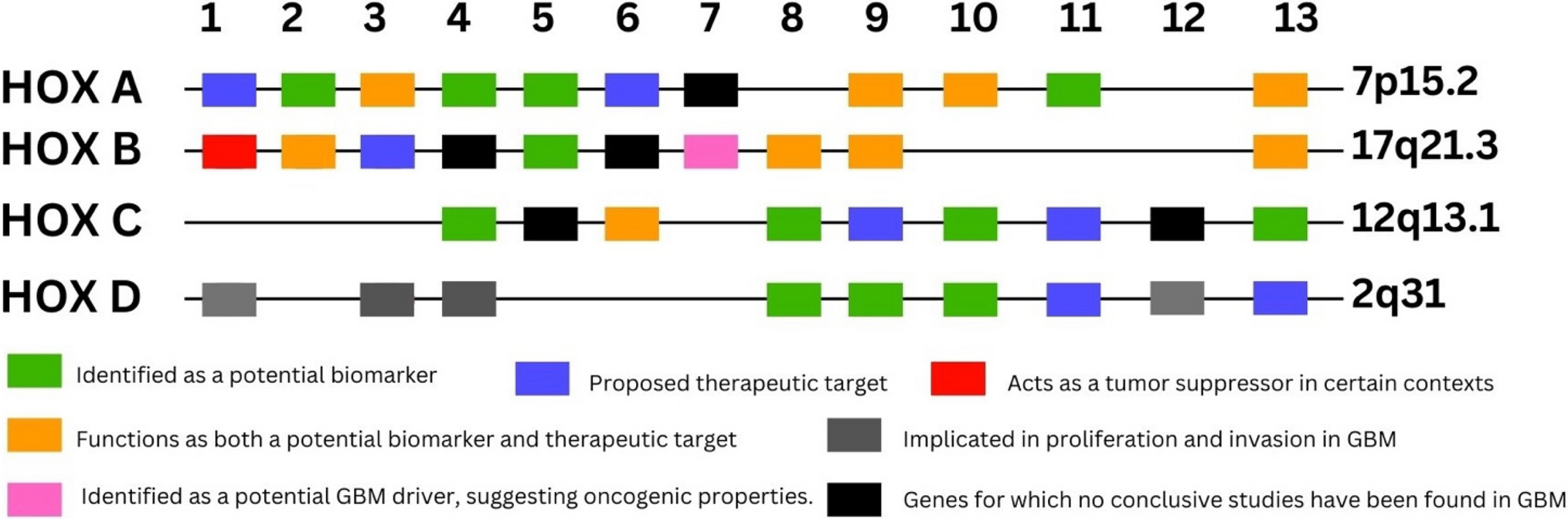
Highlights specific HOX genes (like HOXA9, HOXA10, and HOXD10) that are linked to treatment resistance
-
Explores the connection between HOX gene activity and glioma stem cells, tumor microenvironment, and immune evasion
-
Discusses the implications of HOX genes as biomarkers and therapeutic targets
This is the first narrative review focused specifically on HOX gene dysregulation in GBM.
🚀 Why It Matters
Our findings suggest that HOX genes are more than developmental relics—they may be active participants in the progression and treatment resistance of glioblastoma.
Understanding how and why these genes become dysregulated could lead to:
-
Improved diagnostic tools
-
Personalized treatment strategies
-
New targets for drug development
📖 Behind the Scenes
This project brought together my passion for computational genomics and my fascination with developmental biology. It reminded me how genes that build life can also contribute to its breakdown, depending on the context.
Writing this review wasn’t just an academic task—it was an effort to bridge two fields and encourage researchers to see cancer through a broader genetic lens.
Follow the Topic
-
Discover Oncology

This is a fully open access general oncology journal that aims to provide a unified forum for researchers and clinicians. The journal spans from basic and translational science, to preclinical, clinical, and epidemiology, and welcomes content that interfaces at all levels of cancer research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Single-Cell RNA Sequencing in Cancer Immunotherapy
Cancer immunotherapy is a hot area of current oncology research, with its core focus on activating or enhancing the body's immune system's ability to recognize and kill cancer cells. However, cancer cells possess complex heterogeneity and dynamics, which affect the efficacy of immunotherapy in many ways. Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool in recent years, providing us with an unprecedented insight into the cellular heterogeneity and dynamics within tumors. This technology has revolutionized our understanding of cancer biology, especially in the context of cancer immunotherapy. By enabling researchers to analyze individual cells, scRNA-seq allows them to identify distinct cell populations, track cellular responses to treatments, and discover new therapeutic targets. This collection aims to compile cutting-edge research in this field and explore the various applications of single-cell RNA sequencing in cancer immunotherapy.
This collection will cover the following topics: 1. The latest advances in single-cell RNA sequencing technology in cancer immunotherapy, including research on technology optimization and data interpretation; 2. Using single-cell RNA sequencing to reveal the characteristics of immune cell subgroups in the tumor microenvironment and their interaction mechanisms with cancer cells; 3. Analyzing the molecular basis of immune therapy response and resistance through single-cell RNA sequencing, exploring new biomarkers and therapeutic targets; 4. Combining single-cell RNA sequencing with clinical studies of immunotherapy to assess treatment outcomes, predict patient prognosis, and optimize treatment plans.
Keywords: cancer immunotherapy, single-cell RNA sequencing, therapeutic targets, tumor microenvironment, treatment response
Publishing Model: Open Access
Deadline: Jun 30, 2026
Tumor Microenvironment
The immunological and stromal components of the tumor microenvironment (TME) play important roles in supporting or preventing tumor growth. The interaction of tumor cells and immune cells inside the TME is complex, and it can result in immune suppression, immune evasion, or, in the opposite case, successful immune-mediated tumor elimination. Cancer therapy has increasingly focused on targeting the TME. Immune checkpoint inhibitors (e.g., anti-PD1, anti-CTLA4), CART cell therapy, and vaccinations are all aimed at reactivating the immune system's ability to recognize and destroy tumor cells. Modulating immune component activity is being investigated as a way to counteract the immunosuppressive TME and boost immunotherapy efficacy. This thematic Collection will provide a thorough understanding of the complex role of the tumor microenvironment in the emergence of cancer treatment resistance, as well as innovative strategies for overcoming this challenge and investigating the tumor microenvironment's contribution to personalized medicine.
Keywords: tumor cells, TAMs, TILs, stromal cells, immunotherapy
Publishing Model: Open Access
Deadline: Jun 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in