Fungal “looks” can take your breath away
Published in Microbiology

Invasive fungal infections are an increasingly common complication of modern medicine. Rapid advances in therapies targeting diseases such as cancer, various autoimmune diseases, and extended lifespans of individuals with inborn genetic diseases have increased the number of people susceptible to life-threatening fungal infections. Unfortunately, treatment advances for fungal infections have not quite kept pace with these new emerging infections. Only 3 primary classes of antifungal drugs are in use against the most common mycoses, and the newest, the echinocandins, was originally discovered in the early 1970s. Given the close phylogenetic relationship between fungi and mammals, toxicity is often a major impediment in novel antifungal drug discovery. Another major impediment is a more complete understanding of fungal and host factors essential for respective organism fitness at the site of infection.
To address the latter, our laboratory has focused on defining mechanisms the filamentous fungus (mould) Aspergillus fumigatus utilizes to thrive in vivo at the site of infection. A. fumigatus is the most common causative mould agent of human fungal infections. Previous studies in our laboratory revealed that oxygen becomes surprisingly limited at sites of established infection in the lung. Consequently, we have been utilizing a diverse array of approaches to better understanding how A. fumigatus adapts to a low oxygen environment and how these adaptations impact pathogenesis, virulence, and antifungal drug responses. One recent approach in the laboratory has been to utilize the natural diversity of isolated clinical and environmental strains of A. fumigatus. One striking aspect of this diversity is the stark difference in appearance (“looks”) of A. fumigatus colony biofilms grown in oxygen replete versus oxygen limiting conditions. Strains with increased fitness under low oxygen conditions tend to form more vegetative and aerial hyphae (the individual filaments within a filamentous fungal colony/mycelium). Yet, the genetic factors driving these oxygen dependent morphological changes, and whether they impact pathogenesis or virulence, remain open questions.
To identify genetic factors driving this oxygen dependent morphological heterogeneity, and further understand mechanisms of low oxygen fungal fitness, an experimental evolution experiment was designed utilizing an A. fumigatus strain with reduced low oxygen fitness and reduced vegetative hyphae production. Our goal with this experiment was to let the organism “tell us” what genes and pathways are critical for low oxygen fitness, so we were, in essence, conducting a forward genetics screen. After 20 passages in a controlled low oxygen environment (about 4 months) morphological variants appeared that looked similar to low oxygen fit strains. One variant called EVOL20, displayed this change in colony biofilm morphology but also a significant increase in low oxygen fitness as measured by total fungal biomass production.
In our Nature Microbiology paper (https://www.nature.com/articles/s41564-019-0558-7), we sought to identify and understand the genetic mechanism(s) underlying the change in colony morphology and increase in low oxygen fitness. Once genetic drivers were in hand, we could then address whether these phenotypes played a role in invasive aspergillosis disease progression. Whole genome sequencing revealed a single nucleotide polymorphism (SNP) in a novel unstudied gene located in the sub-telomeric region of chromosome 5. This SNP resulted in a non-synonymous mutation that ultimately turned out to be necessary and sufficient to drive the change in colony biofilm morphology and low oxygen fitness. While open questions remain as to the molecular function of this new gene, the strong phenotypes associated with its loss or over-expression led us to name the gene hypoxia responsive morphology factor A (hrmA). Having isogenic strains with different levels of hrmA expression allowed us to better define the morphology of A. fumigatus colony biofilms as “N-MORPH” or “H-MORPH” and to examine the impact of these morphological changes on the virulence of the respective strains.
As we explored the consequence of increased HrmA expression levels, it became clear that the colony morphology was telling us about the underlying structural arrangement of the hyphae within the fungal biofilm and ultimately something about the hyphal surface of each respective filament. Strains with H-MORPH organize their filaments into arrangements with increased spatial distance. Their adherence to each other and surfaces is reduced and this shapes how the fungal colony biofilm appears on a solid surface. It remains an open question how this morphology impacts low oxygen fitness, but one working hypothesis is that this architecture allows increased oxygen diffusion to deeper parts of the biofilm (see supplemental figure 15 from the paper and adapted here as Figure 1). What ended up being particularly surprising to us was that we could observe this architecture of the fungal biofilm in vivo in a murine model of invasive pulmonary aspergillosis.
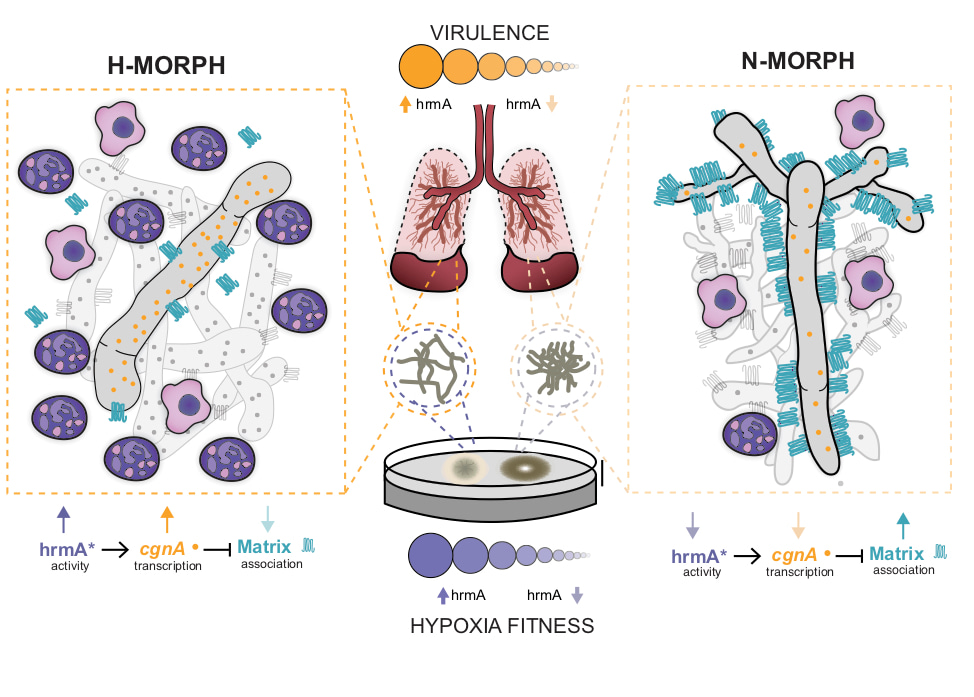
Figure 1. Model for the impact of macroscopic fungal biofilm morphology on Aspergillus fumigatus mediated disease progression. Hypoxic environments found in the A. fumigatus infected lung generate hypoxic-typic morphological features of fungal biofilms including increased furrowing, elevated vegetative hyphal formation, and reduced conidiation. Strains that adapt this morphology have a fitness advantage in low oxygen environments that promotes virulence. Our model predicts that macroscopic fungal morphology (N-MORPH vs. H-MORPH) is an indicator of biofilm architecture and hyphal surface characteristics that impact disease progression. Genetic factors involved in these phenotypes discovered during the course of these studies include the novel genes hrmA and cgnA. Adapted from supplemental figure 15 Kowalski et al. 2019 Nature Microbiology: https://www.nature.com/articles/s41564-019-0558-7.
Strains with “H-MORPH” morphology form less consolidated and more “open” biofilm like structures in the lung and this ultimately correlated with a significant increase in the host inflammatory response to these strains. We hypothesize that the increased disease progression observed with H-MORPH strains stems in part from the altered hyphal surface that impacts exposure or secretion of immune stimulating molecules from the fungus. These molecules are likely to be components of the polysaccharide rich fungal cell wall but we cannot rule out alternations in adhesion like proteins, hydrophobins, or even secreted factors. An intriguing and potentially important observation from the paper is the significant decrease in the attachment of the main polysaccharide component of the Aspergillus fumigatus polysaccharide matrix, galactosaminogalactan (GAG). While loss of GAG did not impact colony biofilm morphology, we cannot rule out that its secretion plays a major role in the disease progression phenotype of H-MORPH strains. Strikingly, it appears that HrmA impacts the expression of a neighboring gene, termed cgnA, whose expression level in EVOL20 impacts the attachment of GAG to the hyphal filaments. As mentioned, significant questions remain regarding the function ofcgnA and hrmA. Their location in the sub-telomeric region of the A. fumigatus chromosome in an apparent gene cluster is intriguing and subject of ongoing research in the laboratory.
In summary, what started out as a relatively straightforward forward genetics screen looking for variants involved in fungal low oxygen fitness led us onto a new research direction examining mechanisms underlying fungal colony biofilm “looks” and their impact on fitness in the context of infection relevant microenvironments. We’ve learned more about genetic factors impacting how filamentous fungi organize themselves into structured biofilms, how oxygen impacts these processes, and how the structure of these biofilms can impact fungal fitness in the context of virulence and disease progression. Ongoing studies continue to probe into the function(s) of hrmA, cgnA and other genes located in the sub-telomeric gene cluster in addition to in vivo studies on the mechanisms of the increased host response to H-MORPH like strains. We’re particularly interested in how oxygen impacts these morphologies and how these mechanisms ultimately affect antifungal drug susceptibility in vivo. While it is still too premature to definitively say that fungal “looks” can be predicative of the host-fungal interaction, we think our findings suggest further research is warranted in this area to better understand fungal fitness in the context of the host immune response and contemporary antifungal therapies. One can speculate that therapies against H-MORPH strains might, in the future, require host targeted immune modulatory agents in addition to contemporary antifungal drugs and/or target fungal specific mechanisms to prevent fungi from taking our breath away.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in