Holobiont Urbanism: sampling urban beehives reveals cities’ metagenomes
Published in Microbiology

Why Holobionts and What Urbanism
For scientists, the term “holobiont” reframes plants and animals as more than isolated individuals. Reaching beyond the notion of individual organisms, holobionts are symbiotic networks consisting of the interactions between hosts (like you, or a tree) and their associated microorganisms. This could include bacteria, viruses, or fungi in the gut, in the soil, or on the train, and how they all mediate and modify each others’ biology.
Cities are no different – from Brooklyn to Sydney to Wuhan. Although cities are primarily built for humans, cars, and commerce, they (like every one of their citizens) – also host trillions of invisible residents whose metabolism and activity define the cities . Our work (begun in 2015) was initially intended to be provocative and exploratory; in the meantime, our collective engagement with SARS-CoV-2 has revealed it to be prescient.
With the global awakening to airborne pathogens, came a sharp interest in where they come from, where they go, and how they travel. We frequently name viruses after their first known engagement with humans, e.g., Ebola is named for the Ebola River in the Congo, the Hanta virus for the Hantaan river South Korea, and so on. With Covid, and the Flu, we have seen the “South African variant” and other attempts to associate pathogens with places. But is it right or useful to associate specific microbes with specific places ?
Beyond pathogens, microbiology as a whole is mostly benevolent and/or essential to human life. Most of it doesn’t get named for places, and much of it doesn’t get named at all. Thus, this work began with the broad inquiry (far beyond concerns about biosecurity) as to whether the broader landscape of microbes have “home cities” or preferred neighborhoods, or even favorite streets in any given city.
There’s extensive research to show that each individual’s microbiome is distinct from anyone else’s, and relatively stable over time. If that’s the case at the scale of the human holobiont, is it also true at the scale of the urban holobiont? Are cities as unique as humans? As stable? Can we distinguish microbial districts that cover a city or a neighborhood, and do they change over time?
In recent years, metrics for invisible factors have revealed distinct zones and districts in any city in the world. Environmental factors that are relevant to health outcomes, like PM2.5, temperature, or allergens sometimes align with political or economic districts along the red lines that define environmental justice areas.
What’s missing in the arrays of real-time urban sensing of the “smart city,” however, is any insight on the local microbial ecosystem. Our interest in discovering and defining urban microbial ecosystems links to some earlier efforts (by study co-authors Chris Mason and Elizabeth Henaff, among others) which led to using swabs to gather surface samples from subways across cities, and then from around the world. This led to the first global map of microbes and antibiotic resistance in urban environments in 2021 from a global Consortium (MetaSUB), but obviously demands intensive costs, coordination, and labor.
At the same time, the last 10 years have also seen the mature deployment of wastewater sampling, around the world. From theoretical beginnings in 2015 to current large-scale commercial deployments (like Biobot) capabilities have emerged to track the microbial landscape for a building, specific urban areas, and even national and international efforts, like GISAID and the U.S. National Wastewater Surveillance System (NWSS). While this has been invaluable in pathogen tracking among other uses (as for SAR-CoV-2) it also takes the specific perspective on microbial presence “downstream” from human interaction.
Our interests, however, diverged from these existing methodologies in two ways. One, we’re interested in what’s beyond pathogen threats: the healthy microbial systems that help us thrive, and the “mundane” ones that have no known relevance to humans, but which may still serve as stable elements of urban ecosystems. Two, we’re looking for the textured identity of the city itself, without the filter and bias embedded in the bycatch of human activity.
Thus, we set out to find a method that allowed us to capture the urban environment itself, at the neighborhood scale, rather than any specific aspect of its infrastructure, or inhabitants.
Sensing the Holobiont
We began work with two designers, Jack Schulze and Timo Arnall, whose work in “Immaterials” (to “illustrate how our interactions with devices and networks are a part of the fabric of everyday urban life”) were an inspiration in how invisible urban forces could be revealed to the human eye. Working together with Schulze and Arnall, as well as some students at MIT, we set out to find a means of applying the same ethos to biological (as opposed to technological) urban dynamics.

This quickly led to close collaboration with study co-authors Elizabeth Henaff and Chris Mason, both at the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine and the WorldQuant Initiative for Quantitative Prediction at Weill Cornell Medicine. The teams began to plan. A Slack group was created. Meetings began. But some key questions remained.
In searching for a means by which to gather the sample from a neighborhood, we had to grapple with the notion of unit volume (the size of the ecosystem of study) and sample fraction (the relative number of elements sampled from the ecosystem), both important factors in the interpretation of microbial community composition.
In 2023, everyone is now familiar with the nasopharyngeal swab as a means of generating a meaningful sample fraction, from the unit volume of a potential Covid-19 patient. But if your unit volume is a city, the scope of a single swab would correspond to a tiny sample fraction.
In searching for a means to gather sufficient sample fraction, we began to consider engaging more directly with ubiquitous living systems that might “do the sampling for us.” We looked at the history of using DNA from mammalian blood in leeches to detect the presence of endangered or near-extinct species in the field. While leeches produce extraordinary aggregated data, they are obviously poorly-suited for cities like New York. There are, however, species that are more common in urban environments, that – like leeches – gather vast quantities of eDNA or microbial cells in the course of their day.
The ideal candidates presented themselves through a series of news articles about the so-called “Red Honey of Brooklyn,” around 2010.

David Selig of Red Hook, Brooklyn. Credit Ozier Muhammad/The New York Times
The “Red Honey” of Brooklyn was found in an urban beekeeper’s hives, whose honey had become a “red concoction more reminiscent of maraschino cherries, or of cough syrup.” The so-called mystery was solved by tracing the path of the bees to a nearby Maraschino cherry factory. In the aggregate of their natural urban foraging, the simple syrup from the factory was now reflected in the honey the bees produced, when they came back home to the hive on the Brooklyn rooftop.
This story was illuminating: the behavior of urban bees might be a viable means of gathering the microbial context in which they forage. The unique structure, physiology and behavior of bees makes them extraordinary collectors, even passively: they gather far more than what they set out to find.
With this insight, we deployed programs and minor adaptations to existing hives, to work with existing urban beekeepers to gather the “bee debris” from their hives. Once samples were collected, the latest techniques in next-generation sequencing, metagenomics, and computational biology were deployed. Specifically, we used a process called “shotgun sequencing” to fragment and sequence all the DNA across all species in a given sample. This enabled a pan-species map of every sample, spanning bee, human, microbial, viral, plant, and any other species’ DNA, creating the first molecular and genetic snapshots of the urban holobiont.
We then metagenomic analysis tools (alignment and assembly methods) to characterize the collected microbial matter, including the “who” was there, in terms of the species, as well as the “what” was occuring biochemically. Since 2015, these methods have been deployed in New York, Venice, Tokyo, Melbourne, and Sydney; many of these are ongoing.
The metagenomic analyses from these hives confirm that neighborhoods have local microbial signatures that are as distinguishable as the microbial signatures of the individuals who live in them. The specifics of these data can be interpretable with respect to hive health and human health through the presence of known markers, or known species.
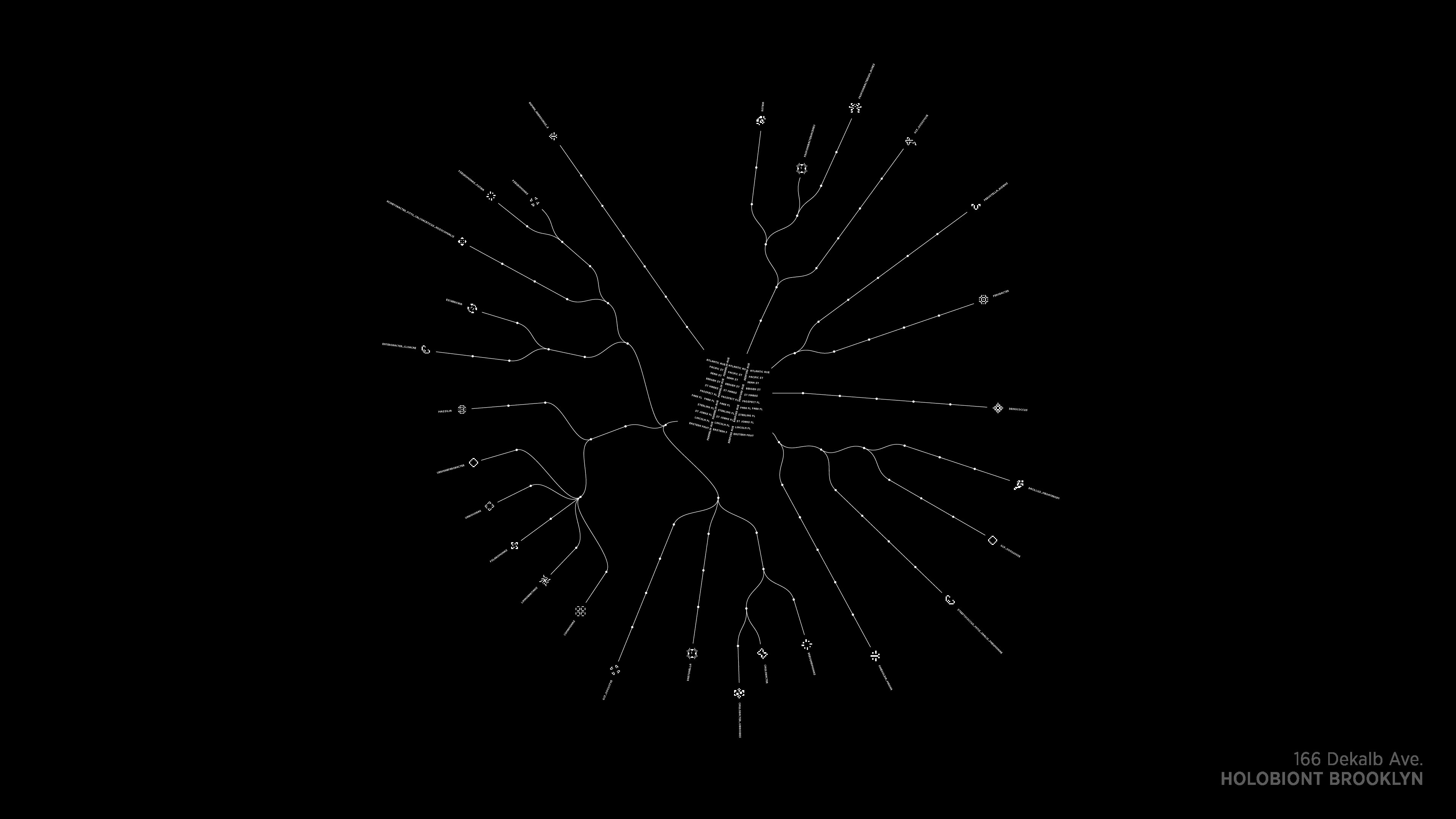
Worth noting, however, is that our goals in the project extended beyond gathering, analyzing and releasing the data. The initial distribution of the ideas, data, and imagery from this work were not in a published paper for scientists to read, but rather in the cultural context of the Venice Architecture Biennale.
The intentions of the work are not only to produce novel findings, but to fuel a novel imagination for the roles of humans, animals, and microbes in the cities we all share.

Holobiont Urbanism exhibit at Palazzo Mora, Venice, Italy during the Venice Architecture Biennale 2016

Detail of data visualization for Holobiont Urbanism exhibit at Palazzo Mora, Venice, Italy during the Venice Architecture Biennale 2016
The method matters in multispecies studies
From the beginning, our use of bees as “citizen scientists” to reveal microbial signatures in cities has been more than a scientific method. It is also a provocation to see our cities from different scales. Beyond the non-human perspectives at the microbial level, we are seeking to engage the world through the lens of interspecies companions all around us. The bees can also stand in for every other interdependent species we live with, without regard for anyone’s specific roles as protagonists.
Our initial inquiry was in considering the urban “holobiont” as a symbiosis between humans and microbes. But by bringing in other inhabitants of the city (the bees) the greater notion of the holobiont comes to life: not a simple bi-directional exchange of value, but rather, the ways that each so-called individual is itself made up of a vast and complex living ecosystem. We set out to reimagine each city we live in as far more than a human metropolis, instead revealing it as a complex and adaptive biological superstructure.
Our goals in this work are to gather, analyze and release data that help to better understand exactly what our cities are. But beyond that, our goal is to reveal that the “empty space” around each one of us is never empty. Our hope is that when you look away from what we show you, you might see the world (or at least your city) quite differently. It is a rich and continual circadian cycle of biology perpetuated by - and pervading - all organisms and all molecules from life; the holobiont.
Follow the Topic
-
Environmental Microbiome

Environmental Microbiome acknowledges the universal presence of microorganisms, which can be found across all environments on Earth, and is seeking submissions addressing the varied facets of environmental and applied microbiological research.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Apple Microbiome
Microbiome and Environmental Microbiome are calling for submissions to our Collection on the Apple Microbiome.
With world apple production estimated at 84 million tons, the microbiome of the apple has significant implications for agriculture, food security, and human health. Understanding the complex interactions between apple plants and their associated microbial communities can lead to improved crop management strategies, enhanced fruit quality and longevity, and sustainable agricultural practices. Recent advances have highlighted the role of specific bacteria and fungi in promoting plant health and resilience against specific pathogens. Moreover, detailed profiling of these microbial communities, revealing their diversity and functional potential facilitate exciting future developments, such as the identification of beneficial microbial consortia for biocontrol and the formulation of tailored probiotic treatments for both plants and humans. By advancing our collective understanding in this area, we can work towards a more sustainable and resilient agricultural system.
Topics of interest include but are not limited to:
-Microbial diversity and function associated with apples
-Effects of soil health and rhizosphere interactions on apple production
-Impact of climate change on the apple microbiome
-Role of the apple microbiome in fruit quality
-Microbiome-driven strategies for disease resistance
This collection is open for submissions from all authors on the condition that the manuscript falls within both the scope of the collection and the journal it is submitted to.
All submissions in this collection undergo the relevant journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief of the relevant journal. As an open access publication, participating journals levy an article processing fee (Microbiome, Environmental Microbiome). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief of the journal where the article is being submitted.
Publishing Model: Open Access
Deadline: May 05, 2026
Harnessing plant microbiomes to improve performance and mechanistic understanding
This is a Cross-Journal Collection with Microbiome, Environmental Microbiome, npj Science of Plants, and npj Biofilms and Microbiomes. Please click here to see the collection page for npj Science of Plants and npj Biofilms and Microbiomes.
Modern agriculture needs to sustainably increase crop productivity while preserving ecosystem health. As soil degradation, climate variability, and diminishing input efficiency continue to threaten agricultural outputs, there is a pressing need to enhance plant performance through ecologically-sound strategies. In this context, plant-associated microbiomes represent a powerful, yet underexploited, resource to improve plant vigor, nutrient acquisition, stress resilience, and overall productivity.
The plant microbiome—comprising bacteria, fungi, and other microorganisms inhabiting the rhizosphere, endosphere, and phyllosphere—plays a fundamental role in shaping plant physiology and development. Increasing evidence demonstrates that beneficial microbes mediate key processes such as nutrient solubilization and uptake, hormonal regulation, photosynthetic efficiency, and systemic resistance to (a)biotic stresses. However, to fully harness these capabilities, a mechanistic understanding of the molecular dialogues and functional traits underpinning plant-microbe interactions is essential.
Recent advances in multi-omics technologies, synthetic biology, and high-throughput functional screening have accelerated our ability to dissect these interactions at molecular, cellular, and system levels. Yet, significant challenges remain in translating these mechanistic insights into robust microbiome-based applications for agriculture. Core knowledge gaps include identifying microbial functions that are conserved across environments and hosts, understanding the signaling networks and metabolic exchanges between partners, and predicting microbiome assembly and stability under field conditions.
This Research Topic welcomes Original Research, Reviews, Perspectives, and Meta-analyses that delve into the functional and mechanistic basis of plant-microbiome interactions. We are particularly interested in contributions that integrate molecular microbiology, systems biology, plant physiology, and computational modeling to unravel the mechanisms by which microbial communities enhance plant performance and/or mechanisms employed by plant hosts to assemble beneficial microbiomes. Studies ranging from controlled experimental systems to applied field trials are encouraged, especially those aiming to bridge the gap between fundamental understanding and translational outcomes such as microbial consortia, engineered strains, or microbiome-informed management practices.
Ultimately, this collection aims to advance our ability to rationally design and apply microbiome-based strategies by deepening our mechanistic insight into how plants select beneficial microbiomes and in turn how microbes shape plant health and productivity.
This collection is open for submissions from all authors on the condition that the manuscript falls within both the scope of the collection and the journal it is submitted to.
All submissions in this collection undergo the relevant journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief of the relevant journal. As an open access publication, participating journals levy an article processing fee (Microbiome, Environmental Microbiome). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief of the journal where the article is being submitted.
Collection policies for Microbiome and Environmental Microbiome:
Please refer to this page. Please only submit to one journal, but note authors have the option to transfer to another participating journal following the editors’ recommendation.
Collection policies for npj Science of Plants and npj Biofilms and Microbiomes:
Please refer to npj's Collection policies page for full details.
Publishing Model: Open Access
Deadline: Jun 01, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in