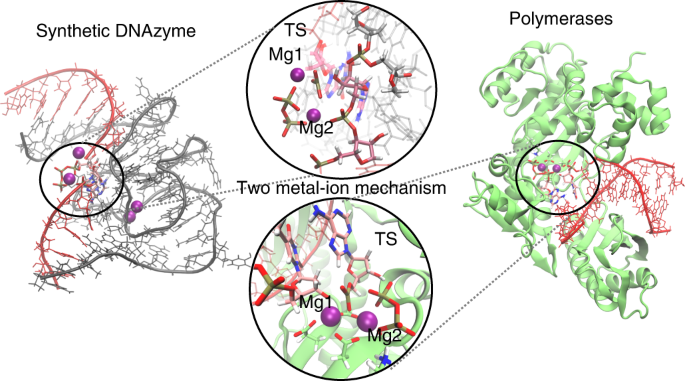
Polymerase-like DNAzyme in action
Published in Chemistry

The role of DNA only being a storage molecule that keeps the genetic information in a stable and irreversible manner has been accepted till recently. In the past decades the discovery of the epigenetic code has broadened the four letters code and defied previous conceptions. In addition, RNA had been solely seen as a genetic carried till its catalytic properties were unveiled in the early 80’s. That gave rise to the term ribozymes and the speculation of RNA being the central molecule of life. Till then, proteins were the only catalysts known in biology. Later, in the 90’s, the first synthetic genetic polymers made of DNA, DNAzymes, were obtained. Since then, numerous DNAzymes have been designed but its mode of action at atomic resolution and whether they were able to adopt complex and compact geometries remained unclear. DNAzymes have attracted much attention because they are easier to synthesise, more stable, less expensive and simpler to use in high throughput in vitro optimisation procedures than proteins or RNAs, and have many applications in biomedicine and biotechnology.
I have always been interested in all of the above because It fascinates me how discoveries in science open new unexpected questions and even defy the existing paradigms. Thus, DNAzymes also were the molecules that gather lots of those interests: structure-function of DNA and RNA, catalysis or theranostics…
During my PhD at U. Valencia I studied the reaction mechanisms of the epigenetic modifying enzymes of DNA (DNMTs), and later on I have been trying to understand how RNA was able to adopt such a broad conformation landscape. I was also trying to predict the folding of catalytically active DNAzymes for theranostics, as there was no structural data available. At that time, the first X-ray structure of a DNAzyme that was crystallised in its post-catalytic state was released. In this X-ray structure no cations appeared near the active site. How does the pre-catalytic state look like? There are no cations involved in catalysis? The high concentration of cations is just needed to acquire the folding?
In this paper, we have investigated at an atomic level both experimentally and computationally how this DNAzyme catalyses the ligation reaction of two RNA strands. Firstly, we made use of a vast number of computational techniques ranging from classical to QM/MM molecular dynamics simulations. We were able to predict where the cations bind in the pre-catalytic state, and we have unambiguously validated these cation binding sites experimentally. We give an atomic description of the reaction mechanism through experiments and computations and characterise the transition state, unveiling important DNA atoms which participate in catalysis and were not identified before. Thus, we were able to analyse the differences and similarities between the catalytic ability of DNA, RNA and the specialised polymerases catalysing the same process. By understanding how DNAzymes work atomistically new and more efficient catalysts may be design.
You can read more about the study in Nature Catalysis.
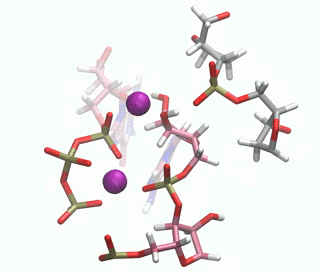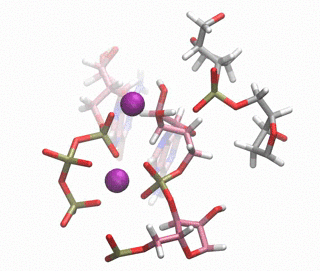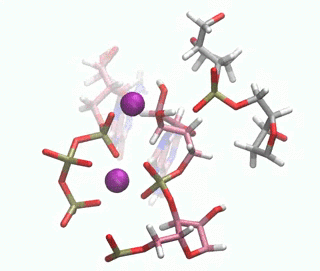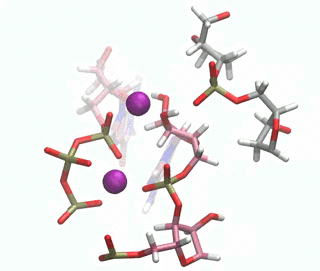
Figure 1. QM/MM-MD of the DNAzyme.
Find us at the Molecular Modelling and Bioinformatics Group led by Modesto Orozco at IRB Barcelona.
Follow the Topic
-
Nature Catalysis

This journal brings together researchers from across all chemistry and related fields, publishing work on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, incorporating both fundamental and applied studies.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in