The Dresden Surgical Anatomy Dataset for abdominal organ segmentation in surgical data science
Published in Research Data

Laparoscopic surgery is performed through small incisions in the abdomen using a thin, tube-like camera and instruments. This surgical technique is used in a variety of medical disciplines including abdominal surgery, gynecology, and urology. Its major advantages are reduced blood loss and trauma, resulting in shorter hospital stays and faster recovery times compared to traditional open surgery. However, laparoscopic surgery can be challenging because it requires surgeons to interpret and navigate complex images in real-time using their anatomical knowledge and medical experience. This is where Artificial Intelligence (AI) could help.
AI-based image recognition methods have the potential to improve safety and efficiency of laparoscopic procedures by automating the recognition of instruments, organs, and anatomical risk and target structures in images and videos. To function reliably, such algorithms need to be trained with large amounts of annotated datasets. However, existing open-source laparoscopic image datasets are limited, with most containing only image-level annotations that allow for determination of the presence or absence of a specific structure in an image, but not its location or appearance.
The Dresden Surgical Anatomy Dataset aims to address this gap. This dataset includes 13,195 laparoscopic images and provides semantic segmentations of eight abdominal organs (colon, liver, pancreas, small intestine, spleen, stomach, ureter, and vesicular glands), the abdominal wall, and two vessel structures (inferior mesenteric artery and intestinal veins). For each anatomical structure, the dataset includes over 1,000 images with pixel-wise segmentations, as well as a multi-organ-segmentation subset with segments for all visible anatomical structures. Additionally, the dataset includes weak annotations of organ presence for every single image.
By leveraging anatomical knowledge, the dataset expands the horizon for surgical data science applications in laparoscopic surgery, which could ultimately lead to a reduction of risks and faster translation into surgical practice. In addition to its value as a research resource, the Dresden Surgical Anatomy Dataset also has the potential to impact clinical practice, as the dataset could be used to train medical professionals in surgical anatomy and laparoscopic image interpretation. This could help to improve the skills and knowledge of the surgical workforce, ultimately leading to better patient care.
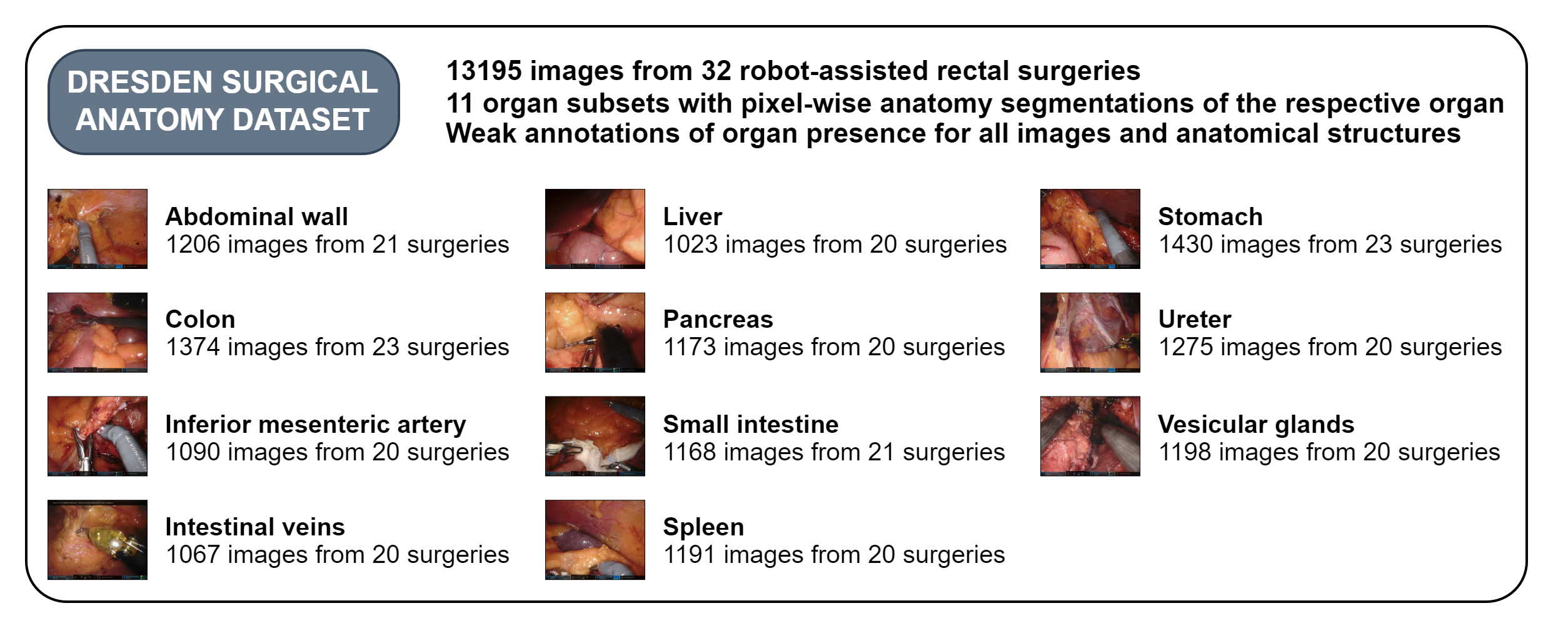
A separate publication provides experimental results on two machine learning algorithms for anatomy segmentation trained on the Dresden Surgical Anatomy Dataset, one for individual structures and one for all structures in parallel. In this study, both models performed on par with human participants in segmentation of the pancreas, a variable-appearing organ that often appears hidden under layers of fat. The models compute fast enough for near-real-time use, underlining the potential to provide valuable assistance in minimally-invasive surgery.
Citations
Carstens M, Rinner FM, Bodenstedt S, Jenke AC, Weitz J, Distler M, Speidel S, Kolbinger FR. The Dresden Surgical Anatomy Dataset for abdominal organ segmentation in surgical data science. Scientific Data 2023. URL: https://www.nature.com/articles/s41597-022-01719-2.
Kolbinger, FR, Rinner FM, Jenke AC, Carstens M, Leger S, Distler M, Weitz J, Speidel S, Bodenstedt S. Better than humans? Machine learning-based anatomy recognition in minimally-invasive abdominal surgery. Preprint at https://www.medrxiv.org/content/10.1101/2022.11.11.22282215v2.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in