A Hitchhiker's Guide to PARIS
Published in Chemistry

The paper in Nature Communications is here: http://go.nature.com/2BVD9jS
A Hitchhiker’s Guide to PARIS
For the past 13 years, our lab has focused on the synthesis, characterization, and application of protein-polymer conjugates. Over the past 13 days we have been struggling over the title for this Blog. Here is the remainder of the list we came up with:
- Why spend days in lab when you can go to PARIS?
- Stuck Purifying Bioconjugates? Let’s Visit PARIS
- Move to PARIS for a simpler way to make bioconjugates
- Your Ultimate Guide to PARIS
- Moving to PARIS? How to settle into new ways of bioconjugate synthesis
- PARIS Adventures
- Explore PARIS
Specifically, we have been interested in using “grafting-from” techniques where we first attach a site to a protein surface from which we can grow polymers by atom-transfer radical polymerization (ATRP). This technique is exquisitely controlled and we have learned how to predict and control where the surface of the protein will be modified. One of the most exciting aspects of this chemistry is that we can make a dense coating on the protein surface. This is the equivalent of growing dense molecular hair, versus constructing a molecular toupee. One of the major drawbacks during the synthesis of these bioconjugates, however, is that it is time-consuming. Bioconjugates are traditionally synthesized in solution and purification via dialysis or diafiltration can take days after each step in the synthesis. In the hands of a skilled bioconjugate scientist it can take a week to make just a few conjugates. We decided to try to redesign the chemistry so that we could significantly decrease synthesis and purification time and make it as simple as possible. The PARIS chemistry that we introduce in this paper is, at its core, simply solid-state protein-ATRP.
The acronym “PARIS” seemed much more appealing than “Protein-ATRP on Reversible Immobilization Supports”. The steps are depicted in Figure 1.
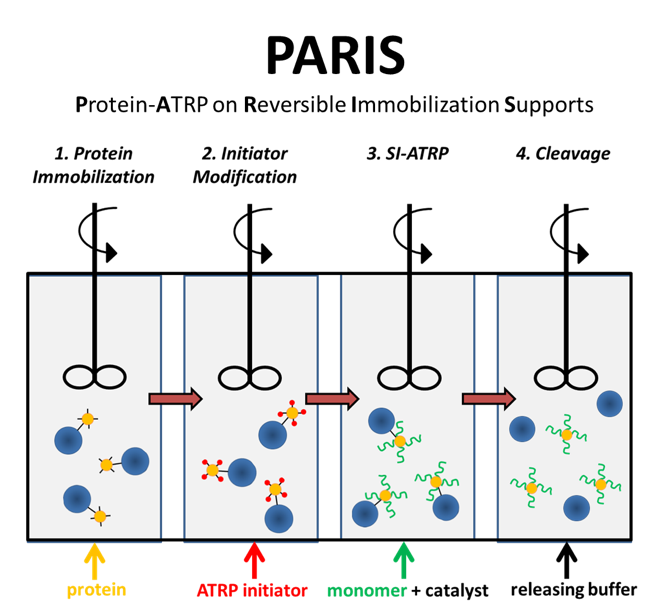
Figure 1. The four steps of PARIS in a flow reactor design.
First, you immobilize protein onto agarose beads. The agarose beads are ~ 45-165 μm whereas proteins are ~2-10 nm. Second, you modify the protein with compounds that can initiate polymerization. After the reaction is complete, you can remove excess initiator by simply filtering the solution since the beads are much larger in size. Third, you add monomer and catalyst of your choice to grow the polymer via ATRP. After filtering the excess monomer, you cleave the protein-polymer conjugate from the beads by reducing pH. After just a couple of hours, you have purified protein-polymer conjugates that we showed have similar activities and stabilities as solution-synthesized bioconjugates.
Another exciting aspect of PARIS is that it allows for automating the synthesis of bioconjugates in a flow reactor. The idea of flowing reactants into a contained vessel and receiving your bioconjugate product at the end is highly appealing for high throughput synthesis, combinatorial chemistry, and scale-up. To test the feasibility of this, we built a basic reactor system containing the beads and sequentially flowed in protein, initiator, monomer+catalyst, and releasing buffer. We are now excitedly ruggedizing the bioconjugate synthesizer and using it to answer previously unanswerable questions. Join us on a journey to PARIS!
Alan Russell, Stefanie Baker, Hiro Murata, Krzysztof Matyjaszewski


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in