Artificial endosymbiosis: Reconstituting a two-billion-year-old marriage of cells
Published in Microbiology

By Jason E. Cournoyer
According to the endosymbiotic theory, modern-day mitochondria and chloroplasts descended from bacteria, once engulfed by a host cell, retained rather than destroyed, and ultimately transformed into organelles through an evolutionary process involving adaptations of the endosymbiont proteome, metabolome, and substantial minimization of the genome. This proposed origin of membranous organelles was sharply panned when advanced by Lynn Margulis in the 1960s, but it has since been vindicated through decades overwhelming phylogenetic and biochemical evidence.
From these studies, we have gained an exquisite understanding of endosymbiosis through its outcomes; for example, we understand that chloroplast genomes were reduced from millions of base pairs in size to a couple hundred thousand base pairs in size. The genes retained in the chloroplast encode tRNAs, rRNAs, and proteins essential for redox regulation, DNA replication, transcription, and translation. We also observe that most of the proteins localized to the chloroplast are encoded by the nuclear genome, synthesized in the cytoplasm and imported to the chloroplast thereafter. Finally, we know that organisms carrying the products of endosymbiosis have themselves been engulfed by other free-living eukaryotes and evolved into a multitude of taxa. While modern-day chloroplasts all originate from one primary endosymbiotic event, the secondary endosymbiotic scenario has occurred many times in evolutionary history.
In contrast to how much we know about endosymbiosis through its outcomes, there is little known about the evolutionary process through which bacteria were established inside host cells and transformed into organelles. The how, in this case, was central to the evolution of complex life forms which forever impacted global ecology in their wake.
In my lab, my coworkers and I have sought to investigate these questions of how by engineering artificial, endosymbiotic life-forms. I believe that without a fossil record or access to naturally-occurring, genetically manipulable endosymbionts, the next best option might be to recapitulate primary endosymbiosis using model organisms. Here, we have endeavored to engineer a model system which can offer insight into: i) what is needed to establish endosymbiosis? ii) how the genomes of the endosymbiont and the host evolve? iii) how did the host and endosymbiont metabolism adapt to endosymbiosis?
Understanding the transformation of bacterial endosymbionts into organelles has been a lasting pursuit. Several studies have reported propagating photosynthetic bacteria, algae and organelles inside eukaryotic cells and embryos. Although these systems show that live, photosynthetic bacteria are capable of transient installation within these unusual environments, the bacteria were not capable of supporting the host bioenergetic functions as chloroplasts do. We envisioned a system whereby genetically tractable cyanobacteria may be engineered to serve essential chloroplast-like functions (such as photosynthesis and fixation of CO2) inside a model eukaryotic host organism. In turn, the cyanobacterium would receive an essential metabolite from the host. In this way, the intracellular bacterium would act not as a benign guest, but as an endosymbiont.
For our experimental design, we were inspired by observations in nature. Freshwater cyanobacteria in the Gloeomargarita lineage are the closest known living relatives to the primary endosymbiotic chloroplasts. Although Gloeomargarita is not genetically tractable, it is related to a basal branch of the Synechococcus-Prochlorococcus clade, which includes several species of fast-growing cyanobacteria that are rendered genetically tractable thanks to pioneering work by Susan Golden and others. Here, we used Synechococcus elongatus PCC 7942 (Syn7942) as a starting strain to engineer endosymbionts. As observed from our studies, this choice proved to be quite significant due to inherent ability of these cyanobacteria to establish bioenergetically necessary endosymbiosis. Because less is known about the nature of the host cell in primary endosymbiosis, we selected a fast-growing, single-celled yeast, Saccharomyces cerevisiae, which is easily genetically manipulated and can be grown under selection conditions.

Figure 1: Some mutant strains of Syn7942 generated in our lab.
Present-day chloroplasts perform a number of functions, including ATP synthesis, assimilation of carbon and nitrogen, and biosynthesis of fatty acids and amino acids. In principle, any of these functions could be leveraged in a platform for establishing endosymbiosis; in fact, the evolutionary history of chloroplasts is obfuscated by genetic adaptations to an extent which precludes easy diagnosis of a single, definitive molecular driver. Bioenergetic considerations, however, have been cited consistently as potential drivers of organelle evolution.
Many genomes of mitochondria, chloroplasts and their extant relatives encode ADP/ATP translocases and transporters. For this reason, we designed our endosymbiotic platform such that the intracellular cyanobacteria supported their hosts on the basis of bioenergetics. Interestingly, we found that wild-type Syn7942 cells fused to yeast mutants transiently supported growth due to inherent abilities of the cyanobacteria to both survive and perform ADP/ATP exchange. To engineer robust endosymbiosis, we further engineered Syn7942 to express a gene encoding an ADP/ATP translocase taken a bacterial endosymbiont of amoebae. These cyanobacteria were introduced into S. cerevisiae mutants that are unable to generate their own ATP under our photosynthetic selection conditions. By sequentially propagating the yeast/cyanobacteria chimera on increasingly stringent selection medium, we were able to propagate yeast/cyanobacteria chimera for 15-20 generations.
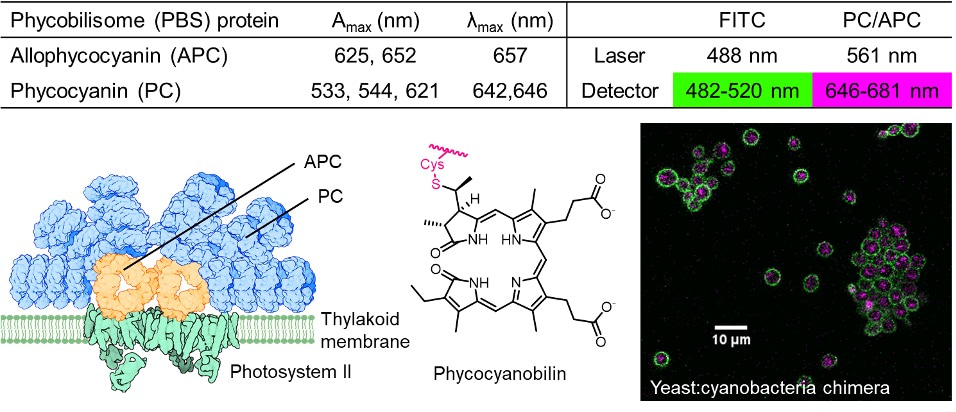
Figure 2: The thylakoid membrane of Syn7942 contains light-harvesting antenna proteins which form the phycobilisome (PBS) complex. These proteins are decorated with chromophores such as phycocyanobilin, permitting easy detection of cyanobacteria by fluorescence microscopy.
My coworkers and I characterized several strains of yeast/cyanobacteria chimera through analysis of total genomic DNA, fluorescence microscopy and TEM. We also showed that the growth of the yeast host cells was supported only under photosynthetic conditions; the chimeras failed to grow when they were deprived of light or exposed to a photosynthesis-blocking herbicide. These experiments supported our notions that the yeast host cells grew thanks to ATP generated by photosynthesis, meaning that the cyanobacteria acted as “synthetic chloroplasts.”
In principle, this genetically tractable endosymbiotic platform can be used to reconstitute and trace the evolutionary trajectory of the chloroplast and other organelles. Further, I believe that these platforms will open up new frontiers in synthetic biology to harness the power of endosymbiosis, just as nature has done for the last two billion years.
References
- Margulis, L. Origin of Eukaryotic Cells; Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth.; Yale University Press: New Haven, 1970.
- Agapakis, C. M.; Niederholtmeyer, H.; Noche, R. R.; Lieberman, T. D.; Megason, S. G.; Way, J. C.; Silver, P. A. Towards a Synthetic Chloroplast. PLoS One 2011, 6 (4), e18877.
- Golden, S. S., Brusslan, J. & Haselkorn, R. [12] Genetic engineering of the cyanobacterial chromosome. Methods in enzymology 153 215–231 (1987).
- Taton, A. et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic acids research 42, e136–e136 (2014).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in