Bacterial nucleoids are more complex than anticipated…
Published in Microbiology
Deinococcus radiodurans is well known for its outstanding radiation resistance – it can withstand very high doses of ionizing radiation and many other DNA damaging agents. In this study, we discovered that D. radiodurans is also particularly well suited for live cell imaging. It is relatively large and a complete cell cycle lasts approximately two hours and is thus neither too slow nor too fast to capture the changes occurring at the cell and nucleoid level during growth and division of the bacteria.
In this study, we wished to address several questions. D. radiodurans has been reported to possess a compact, ring-like nucleoid and these unusual features may contribute to its outstanding radiation-resistant phenotype. So we set out to evaluate the shape and dynamics of its nucleoid as a function of its cell cycle and also wished to use a more quantitative approach to determine the extent of compaction of its nucleoid and compare it to that of the model bacterium, Escherichia coli.
We faced two major difficulties in this study. Imaging small entities such as bacterial nucleoids, which are less than a micron in size, remains a real challenge and we used a combination of spinning-disk confocal microscopy and single-molecule localization microscopy to visualize the nucleoids and probe their conformational dynamics in living cells. However, whilst doing so, we had to overcome a second hurdle, which was that although D. radiodurans bacteria are highly resistant to radiation, they are also particularly sensitive to illumination and thus stopped growing under the microscope! As a result, we had to adapt our illumination schemes and carefully choose our fluorescent probes in order to find experimental conditions that allowed both cell growth and optimal visualization of the cell morphology and the nucleoids.
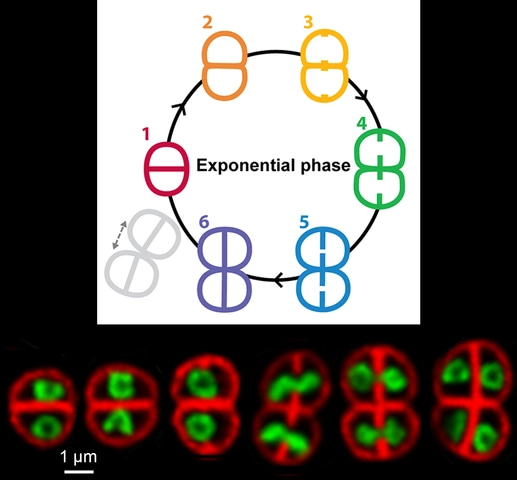
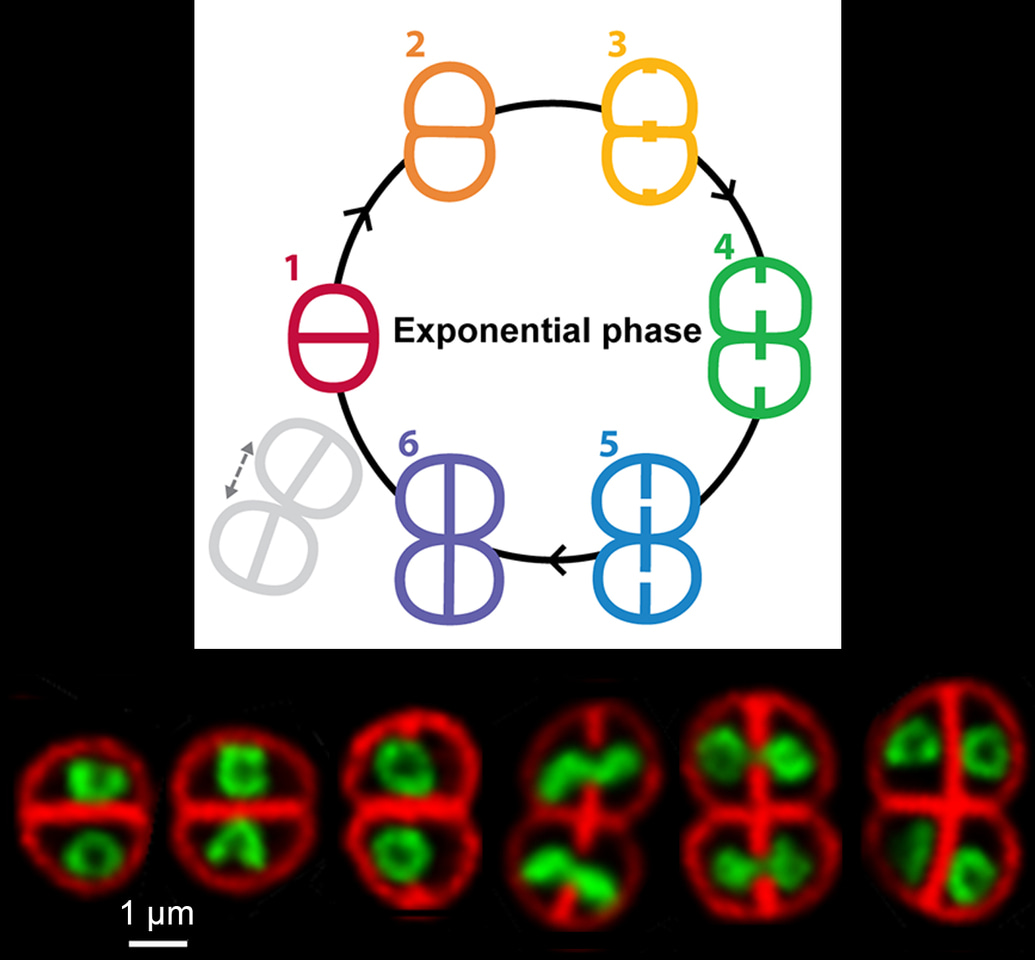
Once all was set, we could get started and we rapidly realized that D. radiodurans nucleoids do not only adopt a toroidal shape, but instead follow a well-defined choreography in which the ring-like structures are just one act of this ballet. Starting from a highly compact conformation just after cytokinesis, the nucleoids then expand to adopt a toroidal shape which progressively opens up into a crescent as the next round of cell division is initiated. As septal growth progresses the crescent-shaped nucleoids stretch along the longest axis of the cell and align orthogonally to the future division axis. At this stage the replicated genomes are pulled into the two daughter cells before septal closure and the newly formed nucleoids are ready to start a new cycle again. Much is known regarding chromosome condensation and segregation in eukaryotes, but so far these processes had never been visualized in bacteria. In this study, we clearly evidenced that bacterial nucleoids are not simple, static structures, but are instead highly complex structures that are tightly coupled to cell cycle progression. We sometimes had trouble believing what we were seeing under the microscope!
We then made use of these beautiful images to determine the dimensions, and hence the volume, of both the cells and their associated nucleoids at the different stages of the cell cycle, in order to evaluate more precisely the level of compaction of these nucleoids. This was a very tedious task, but it provided us with some very interesting data! On average, the nucleoids of exponentially growing cells were found to occupy ~35% of the cell volume in D. radiodurans compared to ~65% in E. coli. In stationary phase cells, this value was found to be even lower with nucleoids occupying less than 20% of the cell volume. These measurements clearly confirmed that the genome of D. radiodurans is indeed highly condensed. Interestingly, unlike other bacteria, D. radiodurans does not possess many nucleoid-associated proteins and its HU protein has been reported to be the major one. In our study, we showed that this highly abundant protein largely colocalizes with the genomic DNA and proposed that its ability to bind only transiently to DNA may contribute to the remarkable plasticity of D. radiodurans nucleoids. But further studies will be needed to determine more precisely the role of this key protein and its potential partners in nucleoid organization in D. radiodurans. It is quite clear that this work only represents the start of the story and in the future we hope to shed light on the mechanisms underlying nucleoid dynamics and its coupling to the cell cycle.
Finally, I wish to point out that this work was largely performed by a highly talented PhD student, Kevin Floc’h, who worked hand-in-hand with our excellent laboratory engineer, Françoise Lacroix. The complementarity between these two individuals was key to the success of this project.
Our full paper: https://rdcu.be/bPqWK
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in