Capturing cells using topological framework nucleic acids
Published in Chemistry
To date, strategies based on multivalent ligands for CTCs capture show limited sensitivity and specificity, typically because of the difficulty in controlling the ligands’ spatial arrangement and stoichiometry.1-3 Given that the receptor−ligand interactions (RLIs) on cell membranes are highly complex and nonlinear, the ability to simultaneously program the spatial arrangement and stoichiometry of the probes therefore represents an approach which can overcome the limitations in CTC capture.
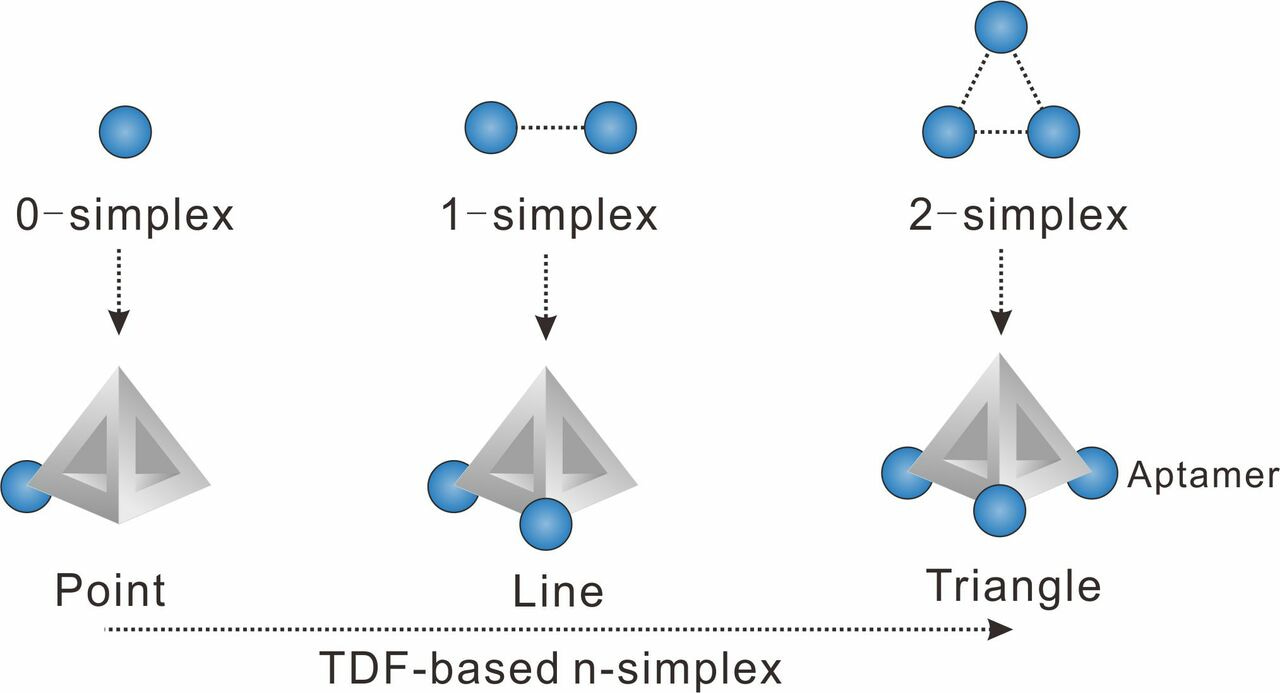
We chose to use tetrahedral DNA frameworks (TDF) to program the spatial arrangement and stoichiometry of probes (here the aptamer of epithelial cell adhesion molecule is used). The TDF-programed probes harboring 1−3 aptamers are termed n-simplexes. The-simplexes, with precisely designed arrangement and the ability to control the stoichiometry of aptamers, possess unique characteristics for topologically programmed aptamers (Figure 1). For example, the 0-simplex harboring one aptamer on the vertex of TDF presents the point-like 0-dimensional distribution of the aptamer. The 1-simplex harboring two aptamers on the vertexes of TDF presents the segment-like 1-dimensional distribution of two aptamers. The 2-simplex harboring three aptamers on the vertexes of TDF presents the triangle-like 2-dimentional distribution of three aptamers.
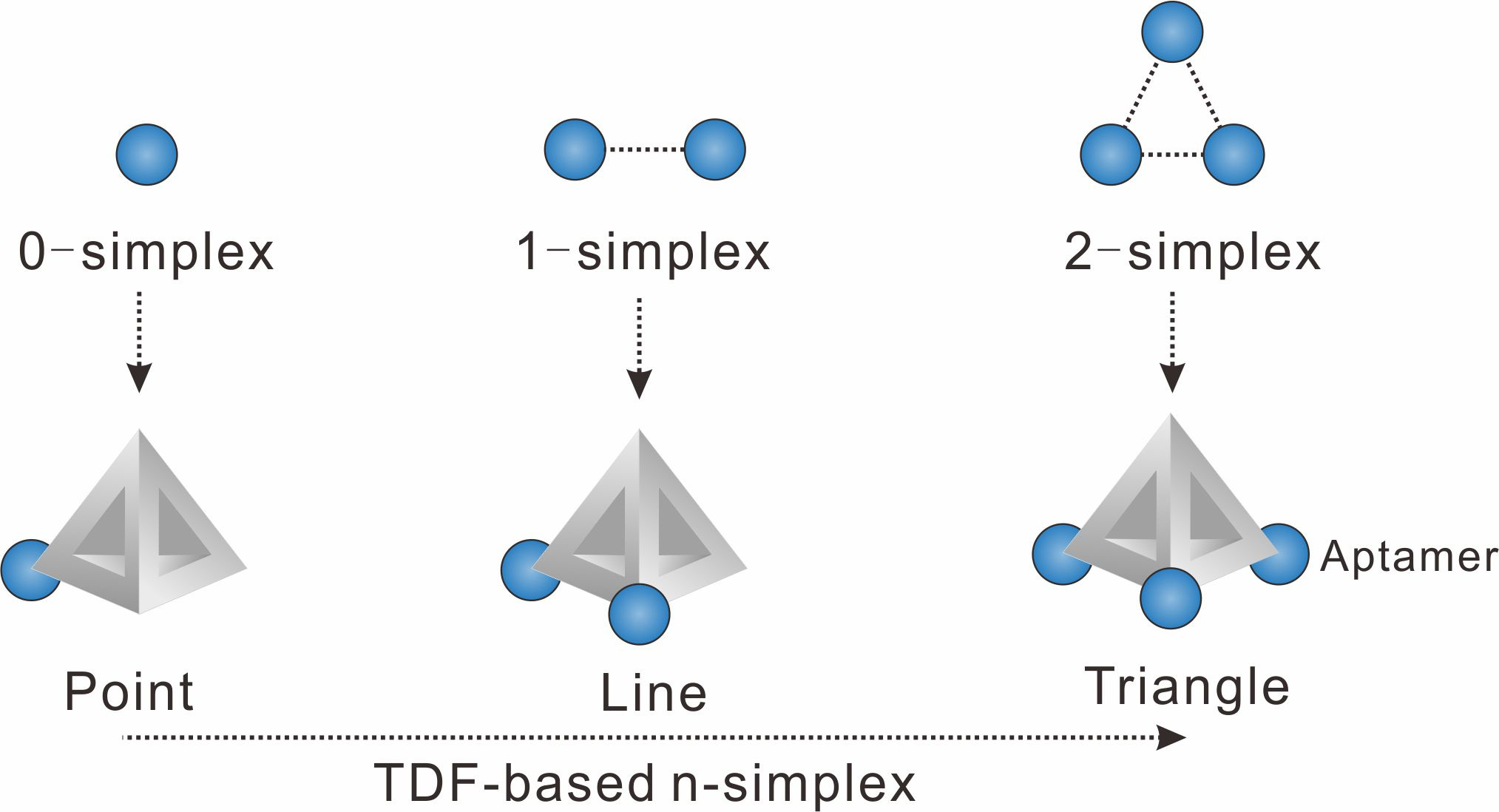
Figure 1. Representations of algebraic topology simplexes (0-simplex (a point), 1-simplex (a segment between two points), and 2-simplex (a triangle)).
The different n-simplexes show different binding properties to tumor cells due to that the algebraic topology affects the RLIs of TDF-based n-simplexes with EpCAM receptors on the cell membrane. Using the rapid binding kinetics, cell membrane adhesion property, and high binding affinity between 2-simplex and tumor cells, TDF-based 2-simplex can be used for tumor cell capture in buffer solution and blood samples with high efficiency.
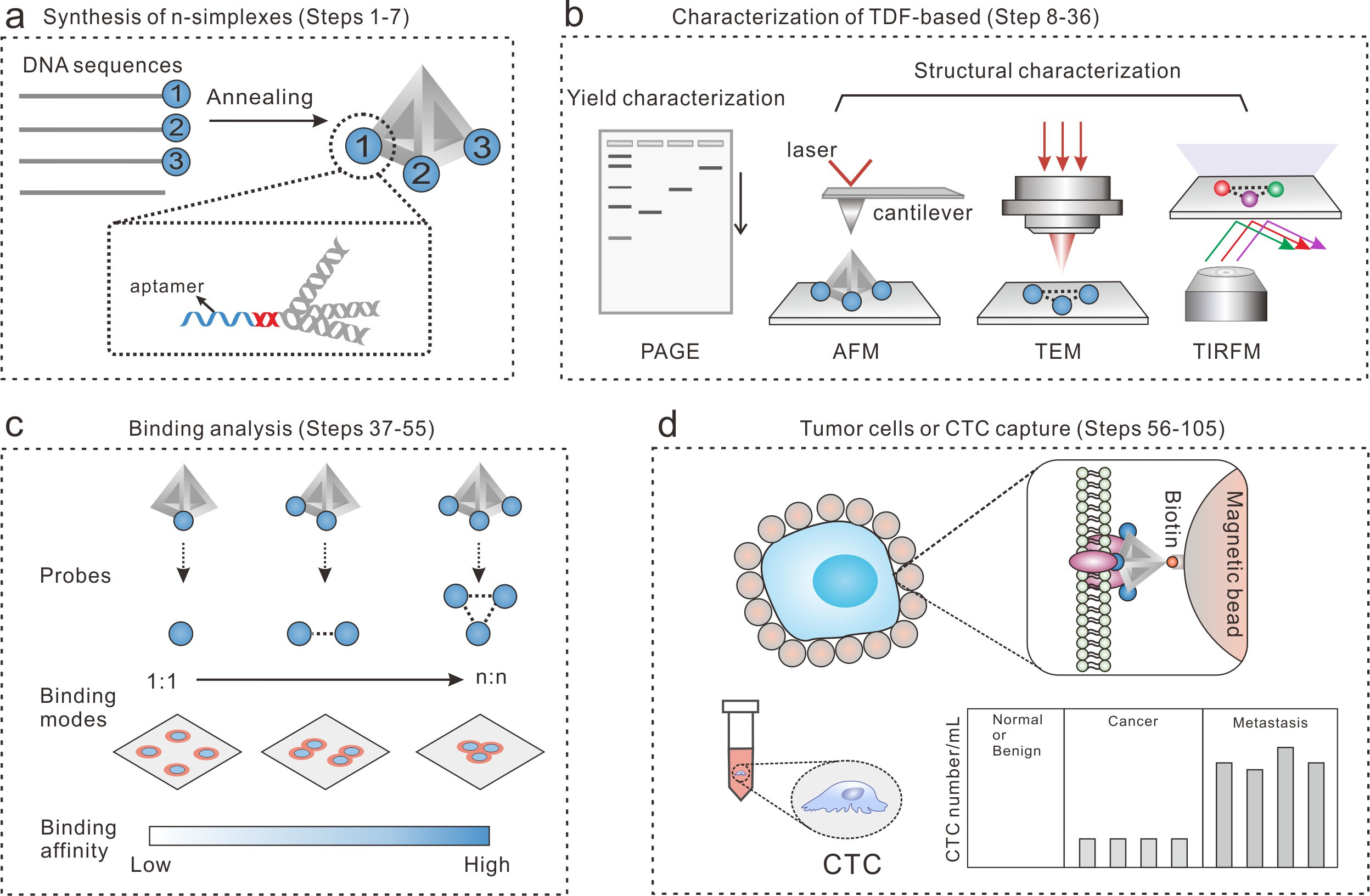
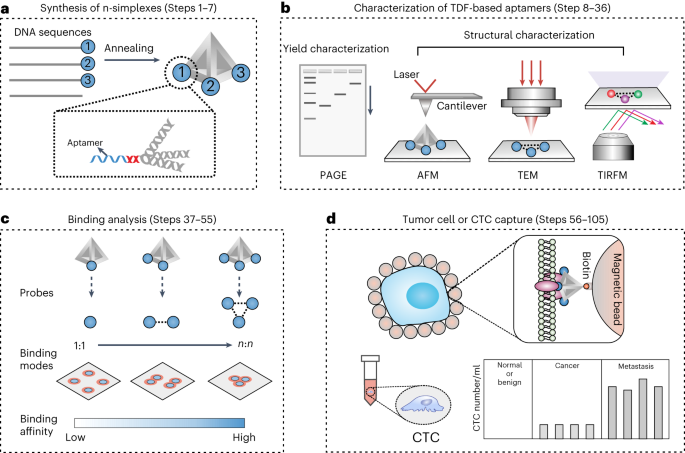
Figure 2. The workflow of the TDF-based n-simplexes for cell capture study. (a) The design and synthesis of TDF-based n-simplexes. (b) The characterization of TDF-based n-simplexes. (c) The binding analysis between TDF-based n-simplexes and receptors on cell membrane. (d) The capture of tumor cells or CTCs.
In our recent article published in Nature Protocols, we summarized the method details for the synthesis and characterization of TDF-based n-simplexes, the binding analysis between n-simplexes and tumor cells, and the capture of tumor cells or CTCs. The present design has potential to be used in a wide variety of areas, including immuno-based CTC detection, microfluidic CTCs capture device, nanostructured surface-based cell capture etc. We expect this new liquid biopsy method to provide new powerful and effective means for cancer diagnostics. And this method has been used in clinical studies for the evaluation of tumor interventional therapy effect.
You can find the online publication from now on via: https://www.nature.com/articles/s41596-023-00943-3
References
1 Song, Y. et al. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2019, 58, 2236-2240.
2 Zhao, W. et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 19626-19631.
- Sheng, W., Chen, T., Tan, W. & Fan, Z.H. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano 2013, 7, 7067-7076.
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in