CDC: A Clustering Algorithm using Local Direction Cenrality
Published in Protocols & Methods

Heterogeneous density and weak connectivity are two common obstacles that have heavy impacts on the accuracy and effectiveness of cluster analysis. Existing methods have difficulty identifying the dense and sparse clusters simultaneously, and separating the weakly connected clusters. In this work, we propose a clustering algorithm named CDC by measuring direction centrality locally, which contributes to handling data with heterogeneous density and weak connectivity. The core idea is to detect the boundary points of clusters firstly, and then connect the internal points within the enclosed cages generated by surrounding boundary points. Specifically, an internal point of clusters tends to be surrounded by its KNNs in all directions, while a boundary point only includes neighboring points within a certain directional range. Taking advantage of this difference, we measure the local centrality by calculating the directional uniformity of KNNs to distinguish internal and boundary points. Hence, CDC can avoid the cross-cluster connections and separate weakly-connected clusters effectively. Meanwhile, it can preserve the completeness of sparse clusters, since it utilizes KNN to search the neighboring points that is irrelevant to the point density.
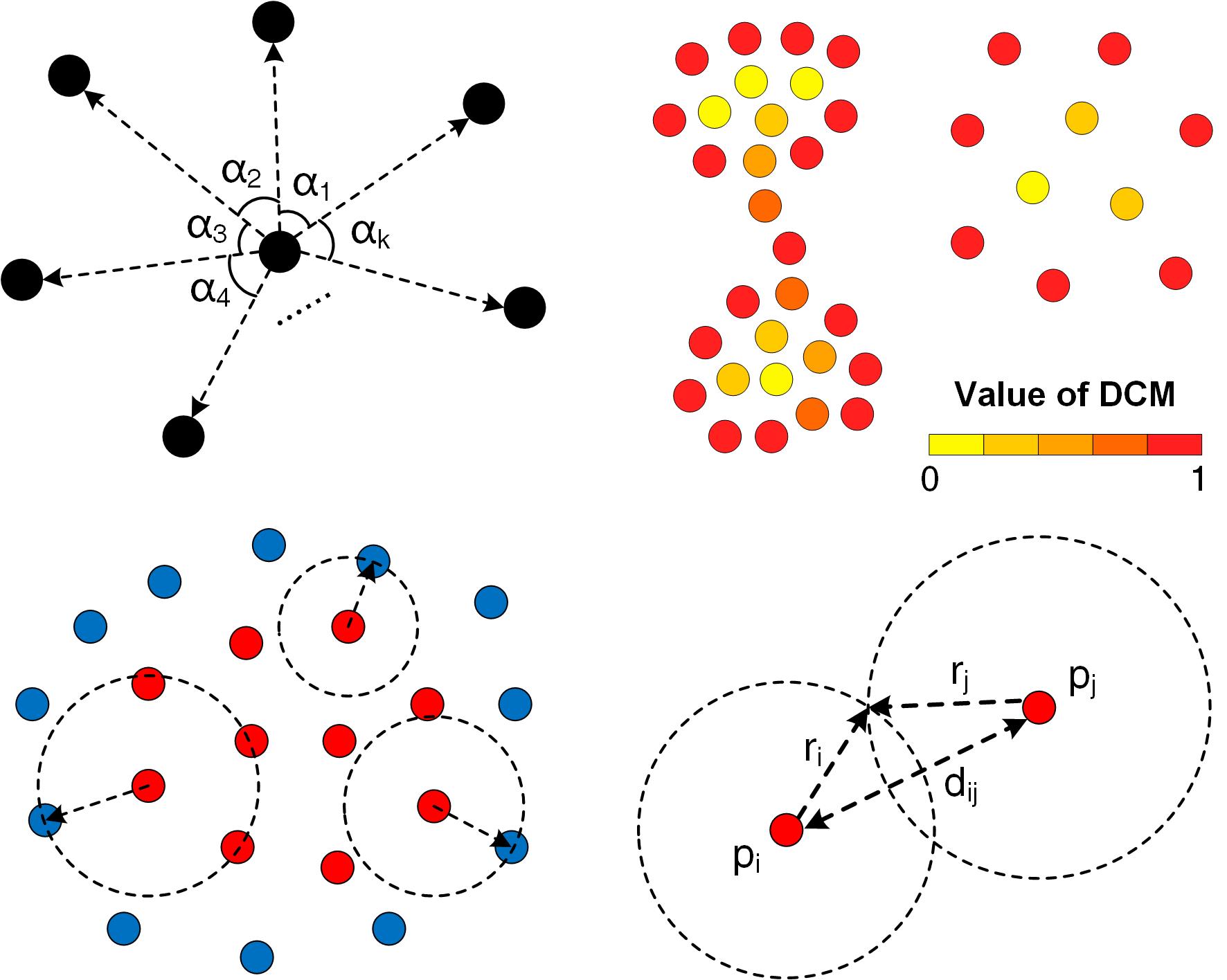 |
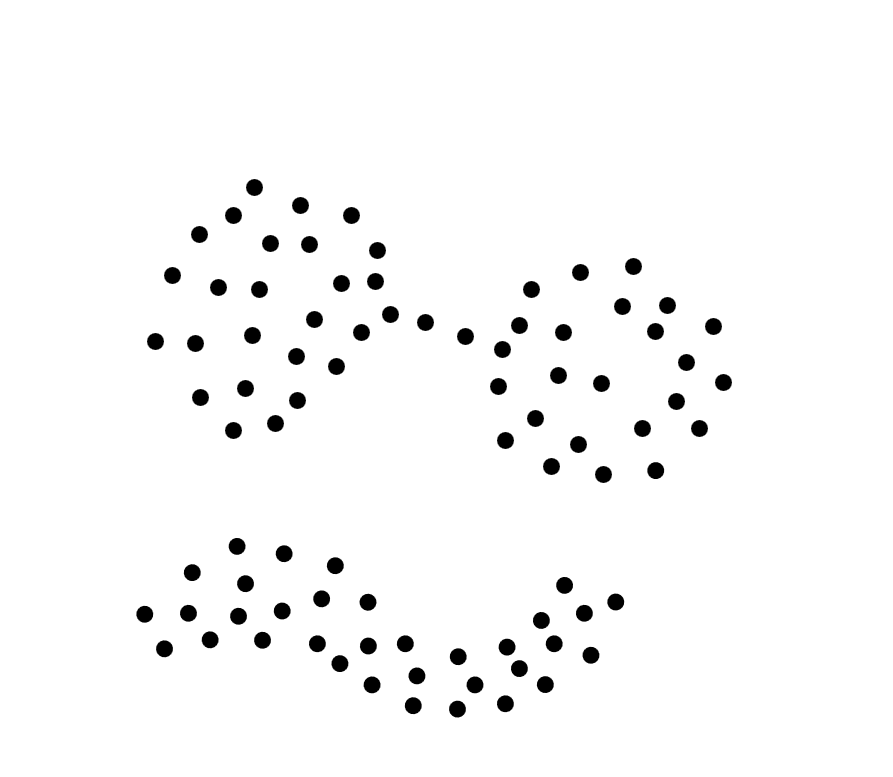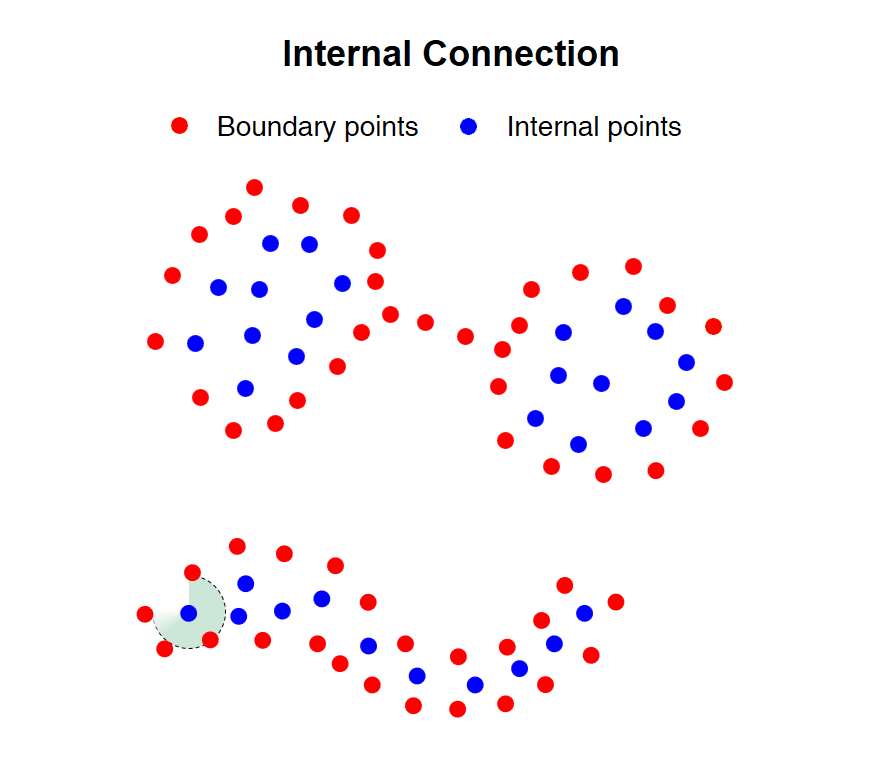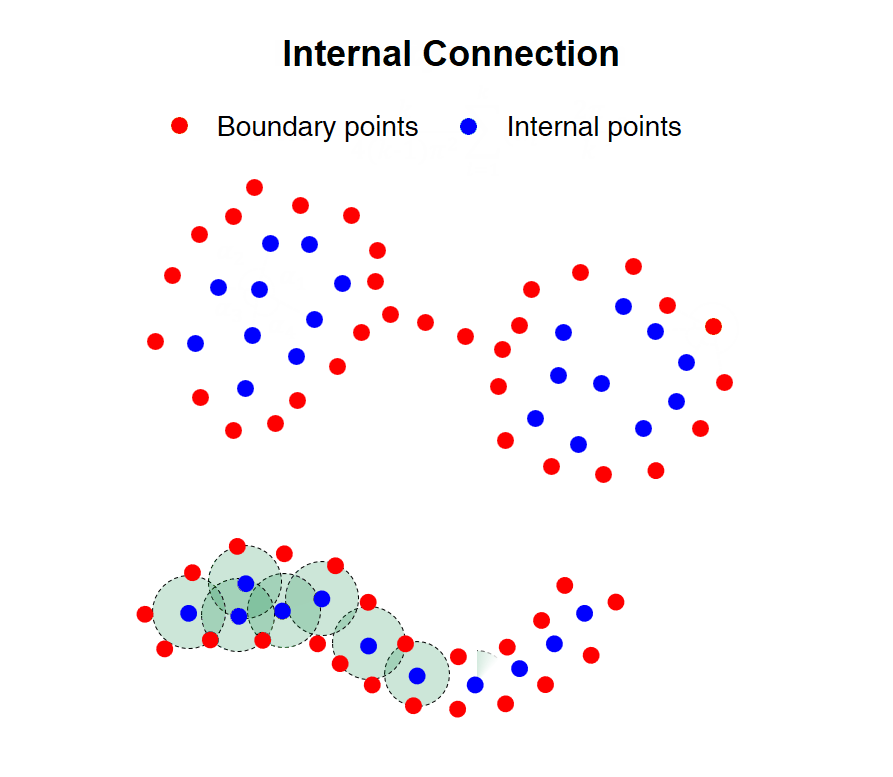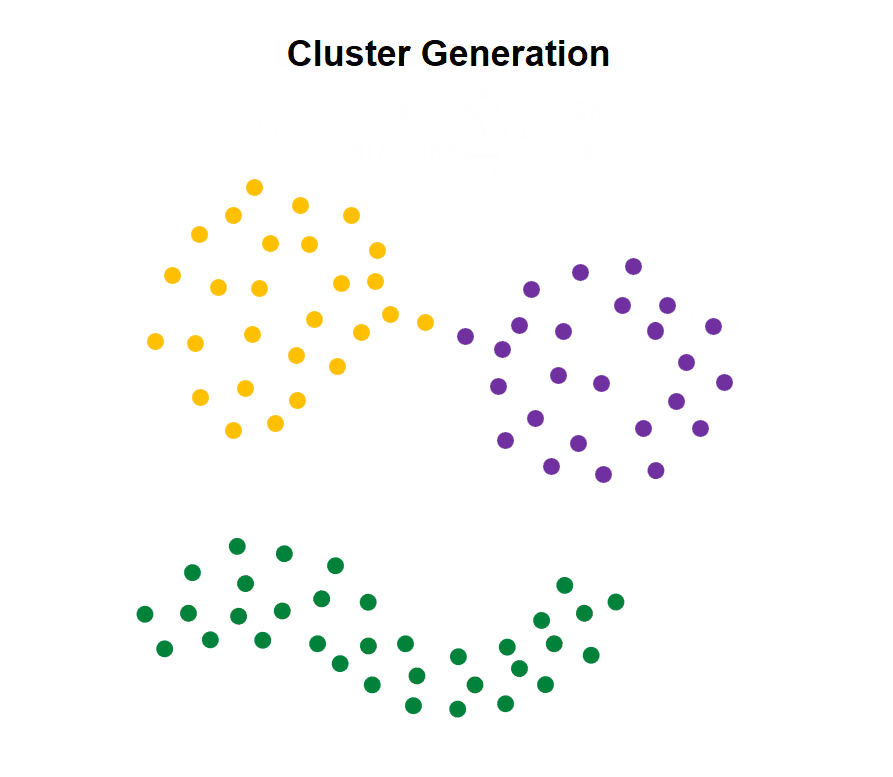 |
Fig. 1 Illustration of CDC algorithmic principle
To validate the effectiveness, we compared CDC with totally 38 specialized and versatile baselines on 47 datasets derived from different fields, including 15 scRNA-seq, two CyTOF, two speaker corpuses, eight UCI, one handwritten image, one face image and 17 synthetic datasets. Results demonstrated that CDC attains superior clustering accuracy and robust outcomes in a time efficient manner, and presented its great potentials in various applications. Moreover, we investigated the dimension expansion and noise elimination methods, analyzed the parameter sensitivity, and designed adaptive methods for parameter settings.
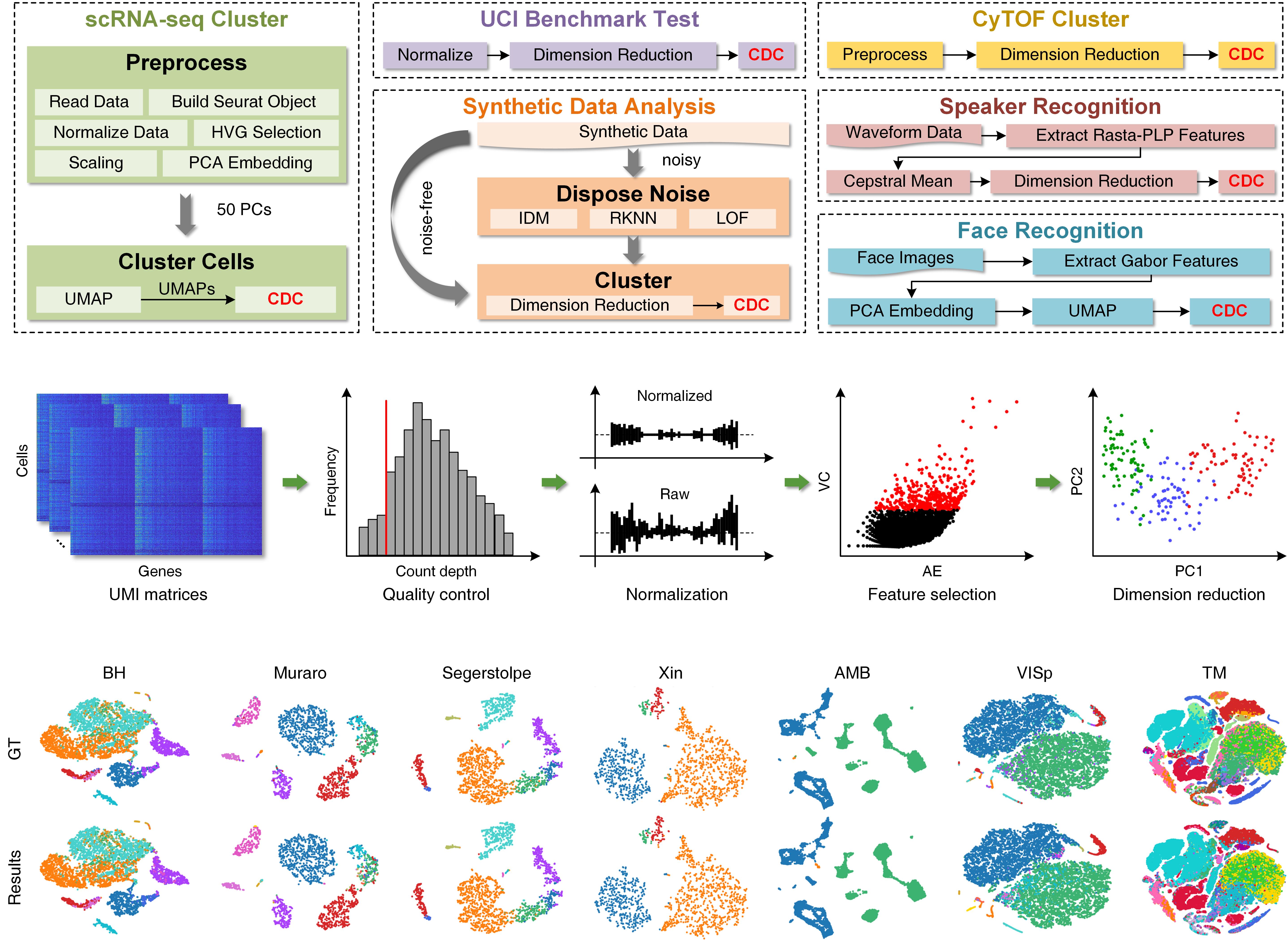
Fig. 2 Six typical applications of CDC, and overview of the standard preprocessing pipeline and clustering results for the identification of cell types from scRNA-seq datasets
CDC is of general significance and has more potentials beyond identifying the cell types, recognizing speaker voices and face images. It can be a promising technique to segment the cell images, explore the spatial living patterns of species, and reveal the aggregation distributions of geographic objectives. However, CDC may be invalid to handle data with manifold structure directly, since the detected boundary points cannot constraint the internal connections in all directions in the feature space. Utilizing dimension reduction techniques such as UMAP to embed the data to a proper dimension can broaden the application of CDC.
The code of CDC in MATLAB, R and Python, and the toolkit with six applications can be downloaded at https://github.com/ZPGuiGroupWhu/ClusteringDirectionCentrality and https://zenodo.org/record/7029720#.YwuFsuxByZw. Digital Object Identifier (DOI) 10.5281/zenodo.7029720.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in