Continuous evolution of base editors, one year on
Published in Bioengineering & Biotechnology
Explore the Research
Continuous evolution of base editors with expanded target compatibility and improved activity - Nature Biotechnology
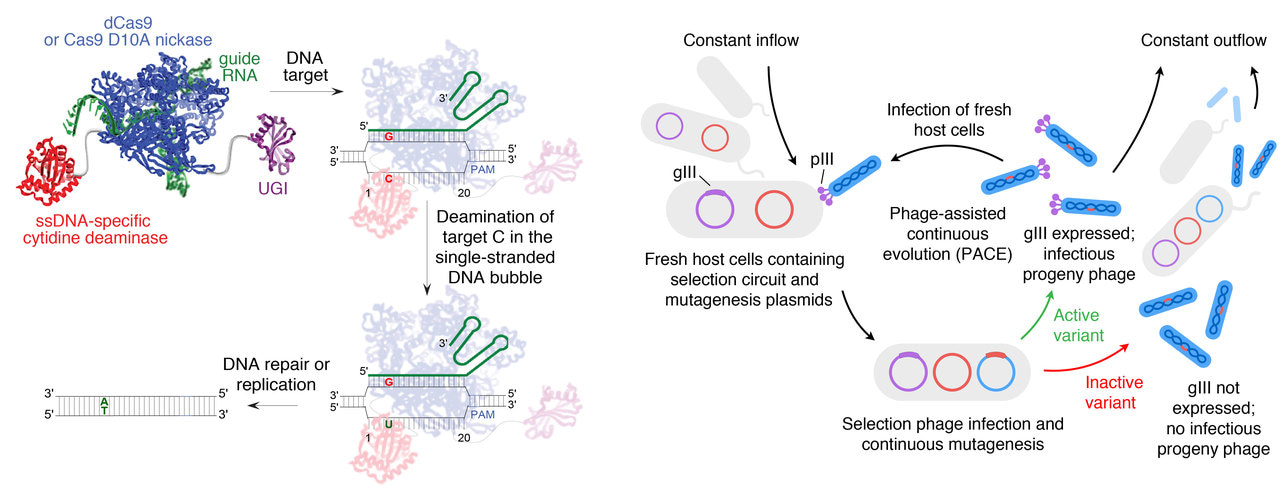
Improved base editors are generated by continuous evolution.
A bit more than a year ago, I wrote here about our work on developing a laboratory evolution method for cytidine base editors (CBE). I said I was most excited about “the work that has yet to be done” – the use of the method we built to take on more challenging goals. I also hoped we’d expand our “picture of how base editing actually works,” and that what we’d accomplished could help.
So what’s happened since then? I was amazed when, less than a year later, my colleagues Dr. Michelle Richter and Kevin Zhao published their evolution of new adenine base editors (ABE) by adapting and building on the CBE phage-assisted continuous evolution (PACE) method.1 ABE is perhaps a more therapeutically impactful target than CBE, since ABE can address more clinically relevant mutations with a cleaner editing profile. On the other hand it was also a far tougher target, since the laboratory-evolved deaminase at the heart of ABE seemed to be much slower than the enzymes with which I started the CBE project. At first, ABE couldn’t activate the easiest PACE circuits we could design. But the existence of a precedent can give you the courage to reach further than before. In fact, by the time Michelle and Kevin were done, they had generated a >500-fold faster adenine base editor, ABE8e. They encountered many of the same hurdles I had, but with the benefit of those earlier experiences (and their own formidable capabilities), they blew through them in record time. This was exactly the multiplier effect I was hoping for when I was grinding through the years-long effort of developing the original base editor selection, unsure if it would ever work!
Besides bringing new therapeutic targets within editing reach, Michelle and Kevin’s work contributed to a huge leap forward in our understanding of base editing, namely a detailed, high-resolution cryo-EM structure and kinetic characterization in collaboration with Prof. Jennifer Doudna’s group showing how ABEs engage with target DNA.2 That study provides something all directed evolution practitioners love to see (though usually we don’t get to) – an analysis of why the mutations that appear during evolution actually help.
Along similar lines, Dr. Mandana Arbab and Max Shen filled in more missing pieces about our evolved base editors from the original paper when they developed the elegant and extremely thorough BE-Hive method, which finally brought big data to base editor characterization.3 Among other things, they revealed sequence context preferences for evolved (and natural) BEs that our hard-won, but ultimately pretty tiny, data set in the original paper couldn’t capture. I highly recommend the BE-Hive paper, and the associated tool, to anyone choosing a BE for a specific application. (By the way, if you’re dealing with paywalls, that paper and everything ever published from David Liu’s lab is available on his group website.) Is base editing still relevant, in light of the brilliant, highly versatile Prime Editing?4 Yes, depending on the target and its context, especially because base editing can still often have higher efficiencies and lower indel levels than prime, though look out for that to change as prime editing follows the tradition of steady improvement!
Where do all these developments leave me personally? For better or worse, I’m mostly out of the base editing world now, so though I’m glad to offer my admiration and encouragement from afar, my own research directions aren’t directly affected. I’m sure I wouldn’t be in my current, fantastic job at Williams College if not for the work we put into the CBE evolution paper – though I had accepted the job before we submitted the paper, the story I got to tell about it during my interview was critical. More broadly, I’m bolstered in approaching my new project – working to turn the absurdly fast-growing bacterium, Vibrio natriegens (Vnat to its friends) into a synthetic biology and directed evolution powerhouse – by the evidence I’ve gotten that long, risky projects can get finished, big investments in methods and platforms can pay off, and a collaborative, cooperative approach to science can make everything work and keep working. When it seems like the pandemic has slowed my new lab’s progress to a crawl, that’s what I try to focus on – and I invite you to keep your focus there too!
- Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
- Lapinaite, A. et al. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science 369, 566–571 (2020).
- Arbab, M. et al. Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell 182, 463–480.e30 (2020).
- Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in