Decorating proteins with chemical modifications using split inteins
Published in Bioengineering & Biotechnology

Galleano I., Khoo K. K. and Pless S. A.
Back in Biology101, we learnt that the structure and function of each protein is primarily determined by its amino acid sequence, which in turn is encoded by the universal genetic code within DNA. Manipulate the DNA sequence, and we can get the cell to generate any protein that we want. Well, not quite, since we would be limited to the 20 amino acid building blocks afforded by the genetic code. Chemists, on the other hand, prefer to synthesize peptides and proteins from scratch, which allows them to introduce non-canonical amino acids (ncAAs) with desired properties and functionalities. However, the size and type of protein we can synthesize this way is still limited.
In recent years, scientists have successfully attempted to reprogram the genetic code, enabling the site-specific introduction of more exotic ncAAs into proteins1. Yet the number and type of residues that can be efficiently introduced is still not limitless and several times, during other projects in the lab, we ended up facing some of these issues2,3. When looking into semi-synthetic avenues to manipulate proteins, we came across split inteins, which offer seemingly endless potential for protein engineering4.
Split inteins are naturally occurring protein domains that essentially behave as connectors to join specific protein segments together in a near traceless manner (Fig 1)4.
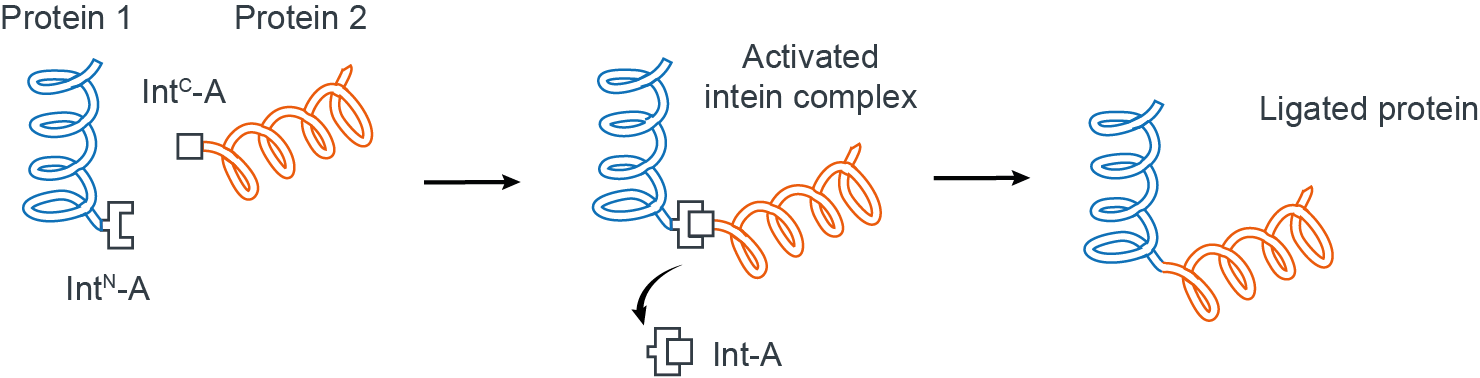
Figure 1. Ligation of two proteins using split inteins.
We thought it should be possible to reconstitute a full-length membrane protein from three distinct fragments using two orthogonal split inteins (Int-A and Int-B) — an approach dubbed tandem protein trans splicing (tPTS). This would allow us to effectively replace a whole segment of the protein with a synthetic peptide, by covalently joining it with the rest of the membrane protein fragments expressed in the cell using split inteins (Fig 2). In theory, this means anything that can be chemically synthesized in the context of the peptide can be incorporated into the protein.
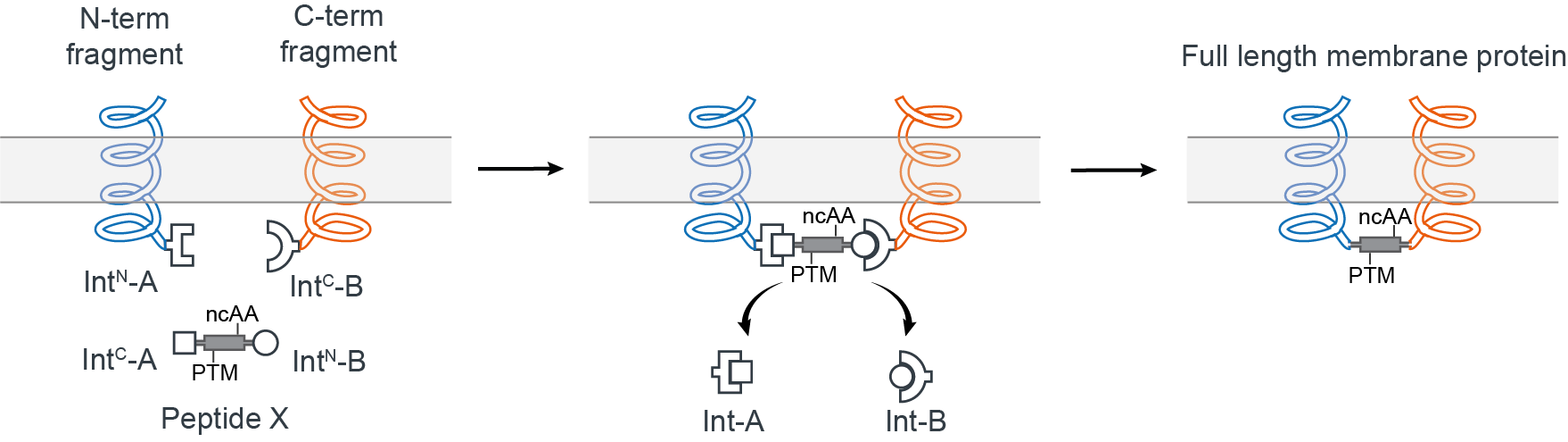
Figure 2. Insertion of a synthetic peptide (Peptide X) into an intracellular linker of a membrane protein.
To showcase some of the possible applications, we used our approach to a) introduce modifications into GFP to manipulate its chromophore properties, b) insert multiple posttranslational modifications (PTMs) into intracellular linkers of the cardiac sodium channel Nav1.5 and c) introduce non-canonical lysine derivatives into the extracellular ATP binding site of the P2X2 receptor (Fig 3)5.
Our work is the first example of reconstitution of full-length proteins by tPTS in eukaryotic cells. The approach has the potential to introduce virtually any ncAAs, PTMs, chemical handles, fluorophores and combination thereof at a chosen site in a protein, thus representing a new tool in the field of protein engineering. Yet it is important to keep in mind that the tPTS yields we experienced in our experiments were typically less than 5%, a value that is likely too low for some assays. However, it is sufficient for investigating proteins that can be studied with highly sensitive methods, such as electrophysiology or imaging.
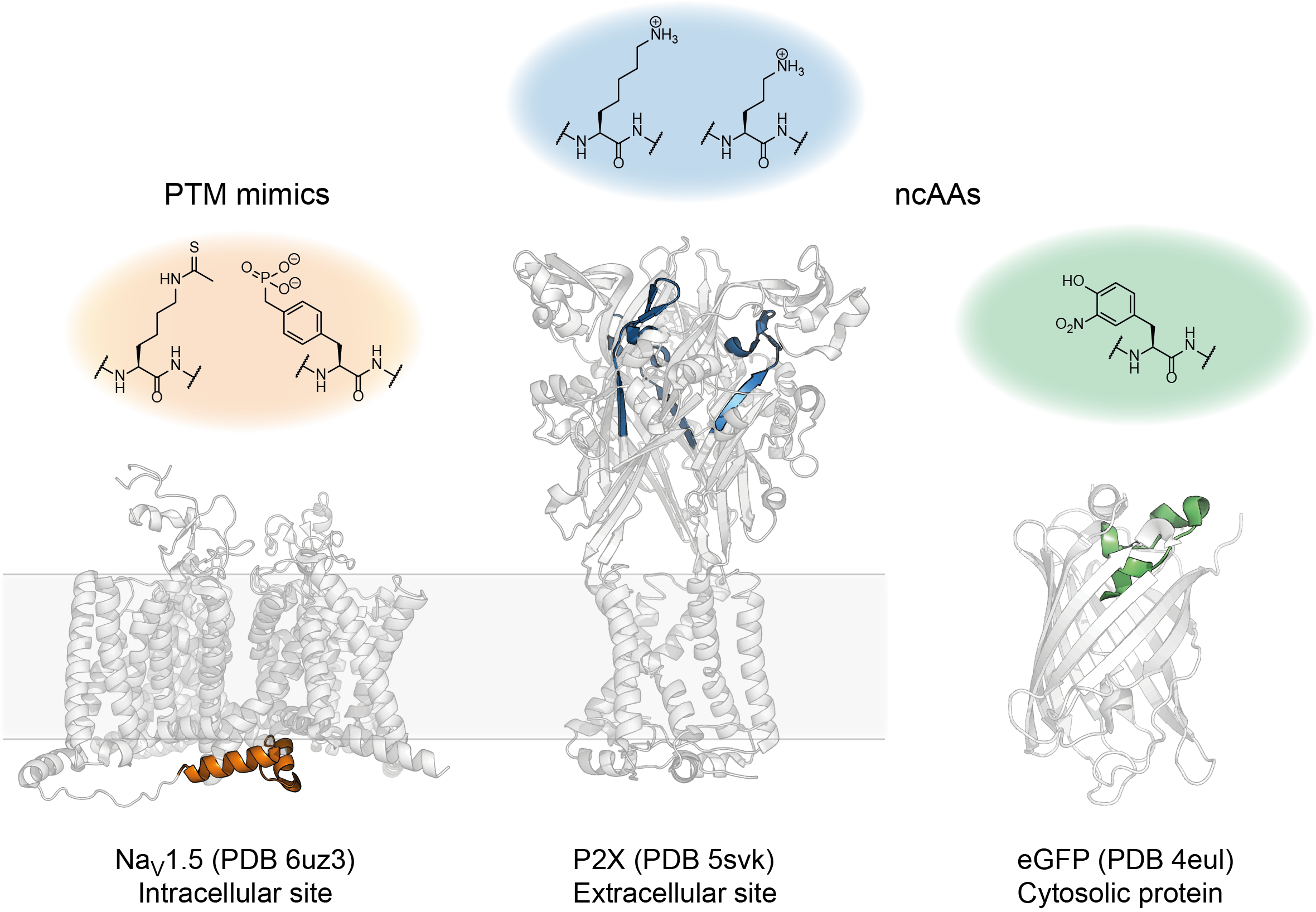
Figure 3. Some of the PTM mimics and ncAAs successfully introduced using tPTS.
Although a powerful tool, there is clearly room for improvement of the technique in the future then. Using more efficient and promiscuous inteins would significantly increase the number of proteins and peptide insertion sites amenable to this approach.
Indeed, with our knowledge of biology and chemistry combined, and a bit of help from split inteins, it seems possible to build and engineer some truly novel proteins in the future.
Read the paper here: Chemical modification of proteins by insertion of synthetic peptides using tandem protein trans-splicing.
1 Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53-60, doi:10.1038/nature24031 (2017).
2 Lynagh, T., Mikhaleva, Y., Colding, J. M., Glover, J. C. & Pless, S. A. Acid-sensing ion channels emerged over 600 Mya and are conserved throughout the deuterostomes. Proc. Natl. Acad. Sci. U. S. A. 115, 8430-8435, doi:10.1073/pnas.1806614115 (2018).
3 Borg, C. B. et al. Mechanism and site of action of big dynorphin on ASIC1a. Proc. Natl. Acad. Sci. U. S. A. 117, 7447-7454, doi:10.1073/pnas.1919323117 (2020).
4 Thompson, R. E. & Muir, T. W. Chemoenzymatic Semisynthesis of Proteins. Chem. Rev., doi:10.1021/acs.chemrev.9b00450 (2019).
5 Khoo, K. K. et al. Chemical modification of proteins by insertion of synthetic peptides using tandem protein trans-splicing. Nat. Commun. 11, 2284, doi:10.1038/s41467-020-16208-6 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in