From the sea to the bench: Marine microeukaryote model systems
Published in Protocols & Methods
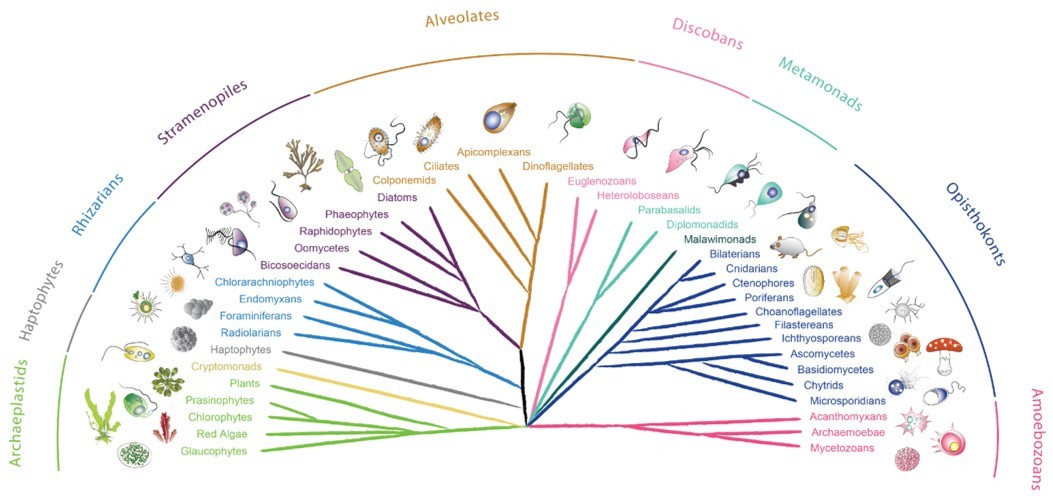
Most molecular and cell biology research is performed on just a few model organisms. The most commonly studied organisms are humans, mice, yeast, worms, the zebrafish, the plant Arabidopsis and the fruit fly. Focusing on this handful of models has been meaningful and productive strategy for understanding organisms in terrestrial and freshwater environments – and during a time when obtaining any DNA sequence data was difficult and expensive. However, there is an enormous diversity of eukaryotic species in the oceans – and these are dominated by single-celled protists. Indeed, most eukaryotic diversity - both terrestrial and marine - is hidden in protists, which do not feature high on the list of model eukaryotes.
Due to their impact on human health, some parasitic protists, such as Trypanosoma brucei and Plasmodium falciparum, the causative agents of sleeping sickness and malaria, are intensely studied. However, considerably less attention has been given to free-living protists. These organisms are present in every ecosystem and have major impact on the habitability of our planet. Even within the protists, the least research has been carried out on marine species, mostly due to the lack of experimental methods – an issue exacerbated by the greater difficulty of cultivating environmentally relevant species. Yet, these are some of the most abundant organisms on planet Earth, and are fundamental to the functioning of oceanic ecosystems. Moreover, they can also be a source of major discoveries based on their unique biology.
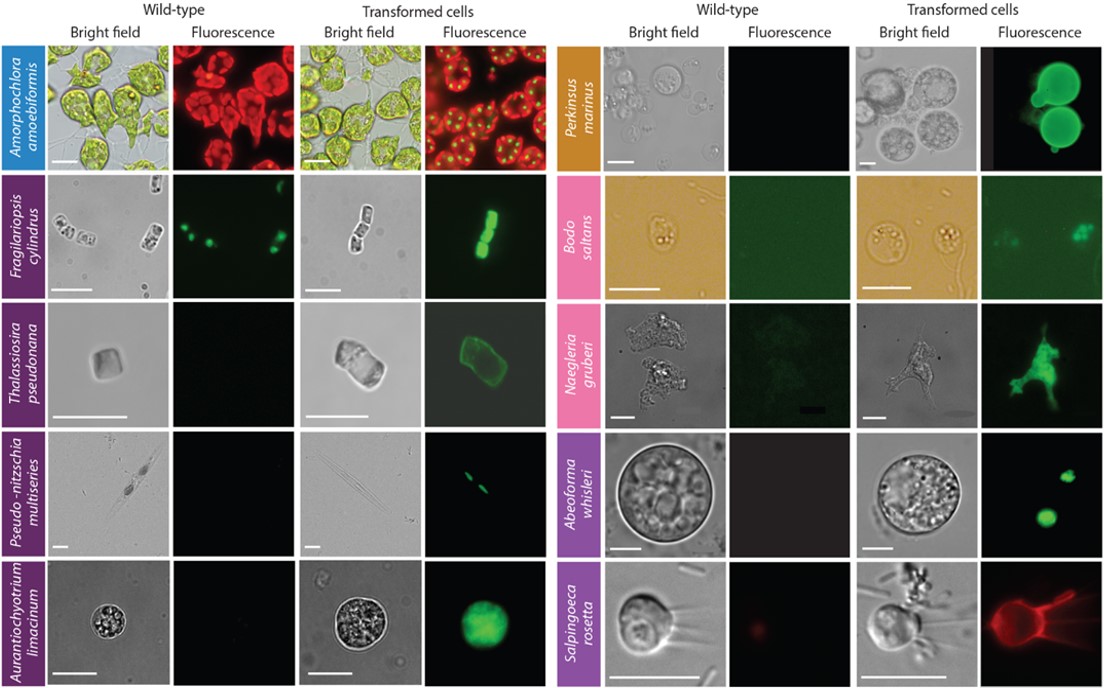
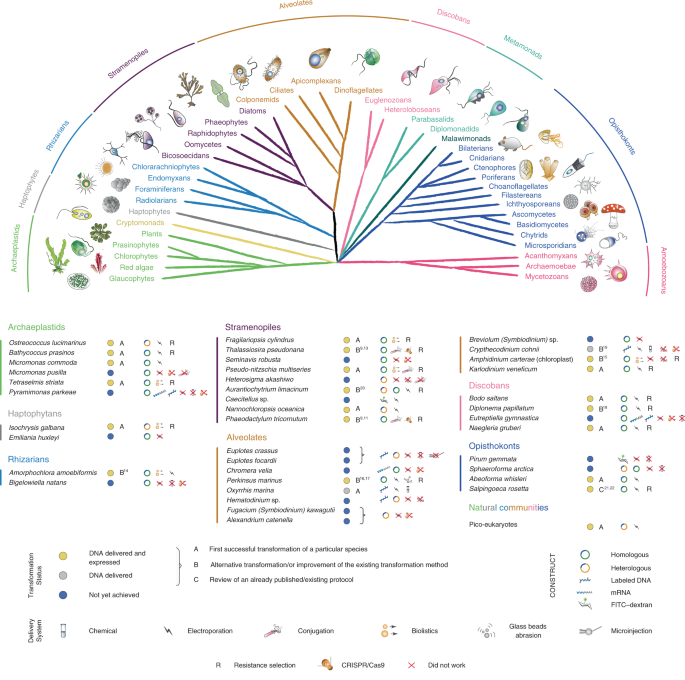
A paper published this month in Nature Methods aims to change the way we view marine protists – and how we can study them. A consortium of 53 laboratories aimed to develop genetic tools in a range of marine protists representing the most significant groups across the eukaryotic tree of life – their collaborative efforts on this topic were supported by the Gordon and Betty Moore Foundation EMS Program of the Marine Microbiology Initiative. As a result, we can now genetically interrogate 13 species for the first time, while advanced genetic tools were developed for a further eight species and we also review an already published EMS transformation protocol in 1 species. Moreover, the paper also contains information about challenges to genetically modify 17 protist species. These unsuccessful approaches are shared to allow others to build on our attempts and to understand which methods did not work, with the hope to encourage alternative DNA delivery methods in the near future.
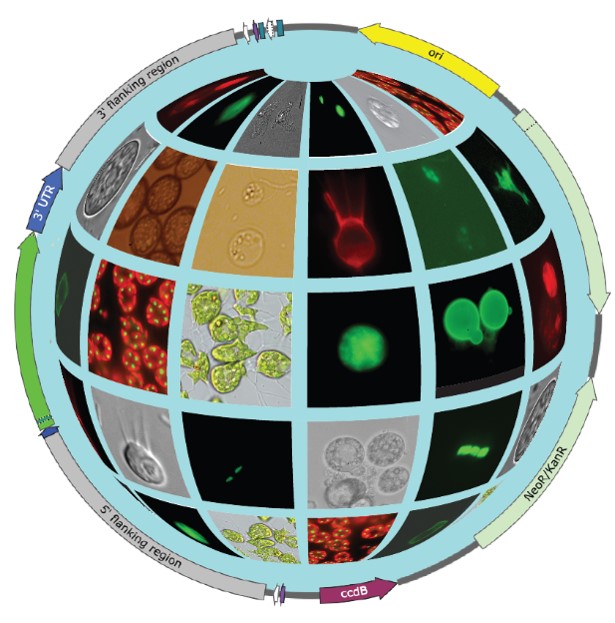
The collaborative effort has been summarized in a ‘Transformation Roadmap’, which provides a path explaining how to create new protist model systems. The first step is usually optimization of cultivation conditions, followed by the identification of suitable selectable markers. As long as at least one method from a wide range of approaches to deliver the exogenous DNA into the cells worked, the chances to establish a new model became high. In the next steps, various strategies were used to integrate the introduced DNA into the host genome. This introduced DNA is transcribed into RNA, and protein products are made. While there is no universally applicable protocol, and a range of conditions have been used, electroporation of linearized plasmids had the highest success rate.
These 22 genetically manipulated marine protist species now provide a springboard for functional studies of the myriad of unique and unstudied genes they carry in their genomes. With more data an unexpected pattern is emerging – namely that numerous processes and structures considered universally present and highly conserved across every eukaryotic cell is instead surprisingly divergent in these protists – challenging our textbook view about the evolution and diversity of eukaryotic life. Collectively the outcomes of this research enable scientists to delve into the secrets of the omnipresent planktonic protists, many of which can be considered the ‘plants of the ocean’ and underpin oceanic food webs and life on Earth.
https://www.nature.com/articles/s41592-020-0796-x
Julius Lukeš, Drahomíra Faktorová, R. Ellen R. Nisbet, José A. Fernández Robledo, Elena Casacuberta, Thomas Mock & Alexandra Z. Worden
For more info see: https://www.moore.org/article-detail?newsUrlName=accelerating-the-development-of-model-systems-for-marine-microbial-ecology-through-collaboration
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in