How to make CRISPR arrays (and what we learned in the process)
Published in Bioengineering & Biotechnology
By Chunyu Liao and Chase Beisel
CRISPR-Cas systems and their Cas proteins have been harnessed for genome editing, gene regulation, imaging, and diagnostics. One unique feature of CRISPR-Cas systems is their inherent ability to conduct multiplexed targeting with CRISPR arrays. CRISPR arrays comprise alternating conserved repeats and spacers that are transcribed into a precursor CRISPR RNA (pre-crRNA) and processed into individual CRISPR RNAs (crRNAs, also generally called gRNAs). While CRISPR arrays offer the most compact means of multiplexing across applications of CRISPR, the re-occurring repeat sequence makes arrays difficult to generate. Specifically, commercial vendors normally reject orders to synthesize arrays as linear dsDNA gene fragments (e.g. see Figure 1A for the output from IDT’s gBlock ordering tool). Although methods for generating sgRNA arrays for Cas9 are widely available, they are only applicable to Cas9 and cannot be extended to CRISPR arrays given their different compositions. In our lab, we work with CRISPR arrays for different studies, but it was always a pain either ordering arrays from commercial vendors or constructing them in-house (https://doi.org/10.1093/nar/gku971, https://doi.org/10.1128/mBio.00928-13). Instead, we yearned for a one-pot assembly scheme to efficiently generate CRISPR arrays for different CRISPR-Cas systems. Moreover, we wanted a modular method for CRISPR array generation that would streamline the construction of libraries of CRISPR arrays, which could largely enhance the power of CRISPR-mediated high through-put screens.
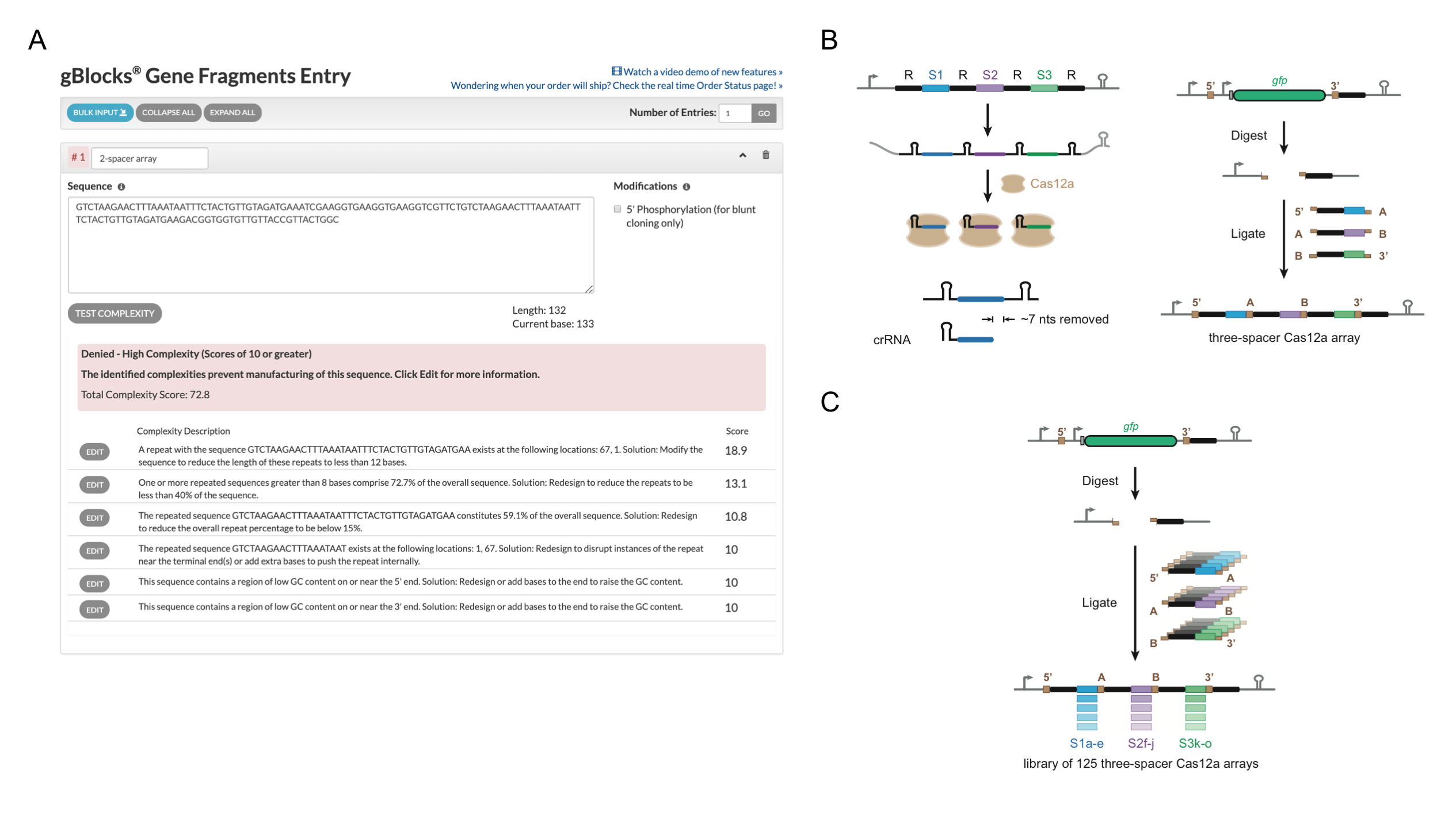
To address this technical challenge, we developed CRATES (CRISPR Assembly through Trimmed Ends of Spacers). As we reported recently in Nature Communications (https://www.nature.com/articles/s41467-019-10747-3), CRATES takes advantage of the portion of crRNA spacers that undergoes trimming as part of crRNA biogenesis. Using defined four nucleotides within the trimmed regions as sticky ends, CRATES allows modular and efficient assembly of short dsDNA bricks, composed of one repeat and one spacer, into large CRISPR arrays and libraries in a single cloning reaction (Figure 1B). As a proof-of-principle demonstration, we made a library of CRISPR array containing 125 different arrays with relatively uniform abundance (Figure 1C). We also confirmed functionality of generated arrays utilized by the single-effector nucleases Cas9, Cas12a, and Cas13a by demonstrating gene regulation and nucleic acid cleavage in prokaryotic and eukaryotic systems. Moreover, we adopted CRATES for generating composite arrays utilized by Cas9 and Cas12a at the same time.
Besides its potential on multiplexing, the fast generation of CRISPR arrays also enables investigation of factors affecting crRNA biogenesis. While testing functionality of a series of CRISPR arrays generated for Cas12a, we noticed lost functionality of a spacer in one configuration in a three-spacer array. The lost functionality was related to the low abundance of the ensuring crRNA, which was further linked to misfolding of a characterized hairpin structure on the 5’ repeat of the spacer on the pre-crRNA. Moreover, we found that the global secondary structure of a transcribed array can help explain the variable abundance of the individual crRNAs for both natural and synthetic arrays, at least in the few instances we interrogated. Another intriguing insight we gained was the biogenesis of an unintended yet functional “extraneous” crRNA processed from the 3’ most repeat in arrays used by Cas12a. Interestingly, naturally occurring terminal repeats for Cas12a often harbor mutations that would be expected to disrupt processing, suggesting that mutations in these terminal repeats appeared under selective pressure to prevent formation of these crRNAs.
As expected, CRATES is becoming widely used in our lab for generating CRISPR arrays for different projects. As one instance, CRATES helped us elucidate how secondary structure flanking CRISPR arrays affects the biogenesis and ensuing targeting activity of crRNAs that guide Cas12a (https://doi.org/10.1080/15476286.2018.1526537). The identified factors relevant to crRNA biogenesis thus could be used for design of synthetic arrays for enhanced multiplexing.
In total, CRATES is capable of facilitating multiplexing with diverse CRISPR nucleases, streamlining combinatorial high through-put screens, and expanding our knowledge of crRNA biogenesis. Given the numerous applications of CRISPR technologies that could benefit from multiplexing with CRISPR arrays, CRATES has the potential to be widely implemented, and we hope the CRISPR community at large can take advantage of this useful method.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in