Isosilybin B: a potential novel therapeutic agent with hepatoprotective, anticancer and antifibrotic properties
Published in Cancer, Cell & Molecular Biology, and Biomedical Research

Since I was a child, I have been fascinated by the unknown in nature. Finding something new always felt a bit like Indiana Jones discovering the Holy Grail - I just hoped mine would be something natural. That curiosity pulled me toward science and, eventually, to a project on milk thistle (Silybum marianum). The seeds of this plant contain silymarin, a mixture of molecules called flavonolignans. The most abundant one is silybin, usually 40–60% of the extract. It was the first to be characterized and naturally became the most studied of all silymarin compounds. But silymarin is more than silybin. Less abundant compounds are hiding in plain sight, for example isosilybin B, typically below 5%. Earlier papers hinted that isosilybin B might have selective anticancer properties, sometimes outperforming silybin. That was the spark for our work: to compare isosilybin B (IB), silybin (SB), and whole silymarin (SM) head-to-head in liver tumor cells (Hepa1-6, HepG2) and non-tumor hepatocytes (AML12).
What we found?
-
A strong killer of cancer cells
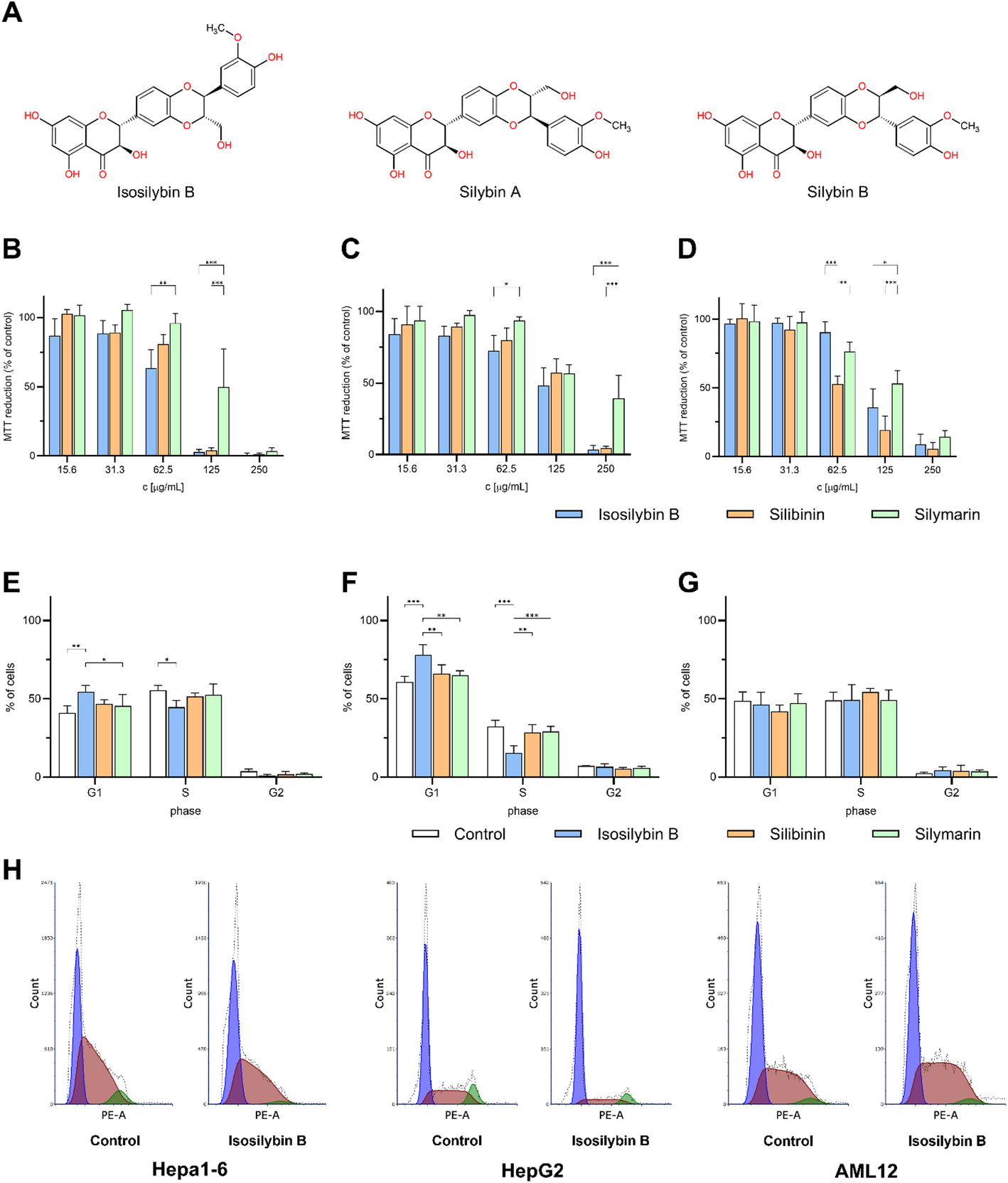
IB was more toxic to tumor cells than SB or SM, while being gentler on healthy hepatocytes. That’s the kind of split we want in an anticancer lead: tough on cancer, kinder to healthy tissue. -
Tumor-specific “arrest”
At a non-toxic concentration, IB arrested the cell cycle of Hepa1-6 and HepG2 cells but left AML12 unchanged. Under the same conditions, SB/SM didn’t show this tumor-selective effect. Mechanistically, this suggests that IB is “wired” into tumor cell biology in a different way. -
Antifibrotic signals went down
When we stimulated cells with TGF-β1 to mimic a profibrotic environment, IB lowered mRNA levels of Acta2 (α-SMA) and Col1a1 (collagen I), in some cases more strongly than SB or SM. At the protein level (fibronectin released into the medium), IB’s effect looked weaker at a single 24 h snapshot but was dose-dependent, hinting that timing and kinetics matter. -
The antioxidant paradox
In a simple chemical DPPH test, the ranking was SM > SB > IB (IB the weakest antioxidant; IC₅₀ ≈ 500 μg/mL). Yet IB produced the largest drop in extracellular ALT (a hepatocyte injury marker) at a non-toxic concentration. The take-home: IB’s hepatoprotection isn’t just “antioxidant”, and is more likely tied to its antifibrotic effects and other cellular processes that a tube test can’t capture.
Why this matters?
Liver disease often involves fibrosis and, for many patients, the looming risk of hepatocellular carcinoma. A molecule that is tough on tumor cells, gentler on healthy hepatocytes, and dampens profibrotic signaling could be valuable across that spectrum. From early liver injury to cancer risk. For a constituent present at only a fraction of silybin’s abundance, IB showed a surprising amount of promise.
There’s also a broader lesson: in mixtures like silymarin, the most abundant compound is not always the most important. Minor constituents can have different 3D shapes (stereochemistry) and different pharmacokinetics. Those small differences can translate into very different biology. IB and SB, for example, have the same atoms arranged differently.
The part you won’t see in the manuscript
At the start, we debated: Is it worth the time and cost to chase a minor constituent? IB is harder to source and more expensive than SB. Early antioxidant runs made IB look underwhelming; they forced us to ask if we were measuring the right thing. The turning point came with the cytotoxicity assays. That was the moment we realized we were seeing a molecule that might truly matter for patients.
What this could change
If IB’s profile holds up in animal studies and, eventually, in clinical settings, it could push milk-thistle research beyond silybin. For patients, the ideal would be a dual-action agent, one that slows tumor growth and eases fibrotic stress in the liver, without punishing healthy hepatocytes. IB might be one such candidate. Even if IB itself is not the endpoint, the concept that lesser-known flavonolignans may outperform the “star” is an important shift in thinking.
What’s next
These are in vitro results, so the next step is in vivo (mouse models) to see whether the tumor-selective, antifibrotic, ALT-lowering profile appears in a living liver. In parallel, we’re exploring delivery strategies, for example, nano-enabled formulations, that could help IB reach the right cells and persist at useful concentrations. We also want to learn more about its mechanism (which pathways trigger the tumor-specific G1 arrest?) and pharmacokinetics (how IB moves and clears in vivo). Finally, we’re curious about combination strategies, can IB work synergistically with existing antifibrotic or anticancer agents?
If those pieces come together, IB could help move silymarin research beyond silybin, and remind us that the “quiet” molecules in a mixture can sometimes carry the loudest signal. While our study focused on milk thistle, the same principle almost certainly applies more broadly; in many medicinal plants, the “minor” compounds may hold major, overlooked therapeutic potential.
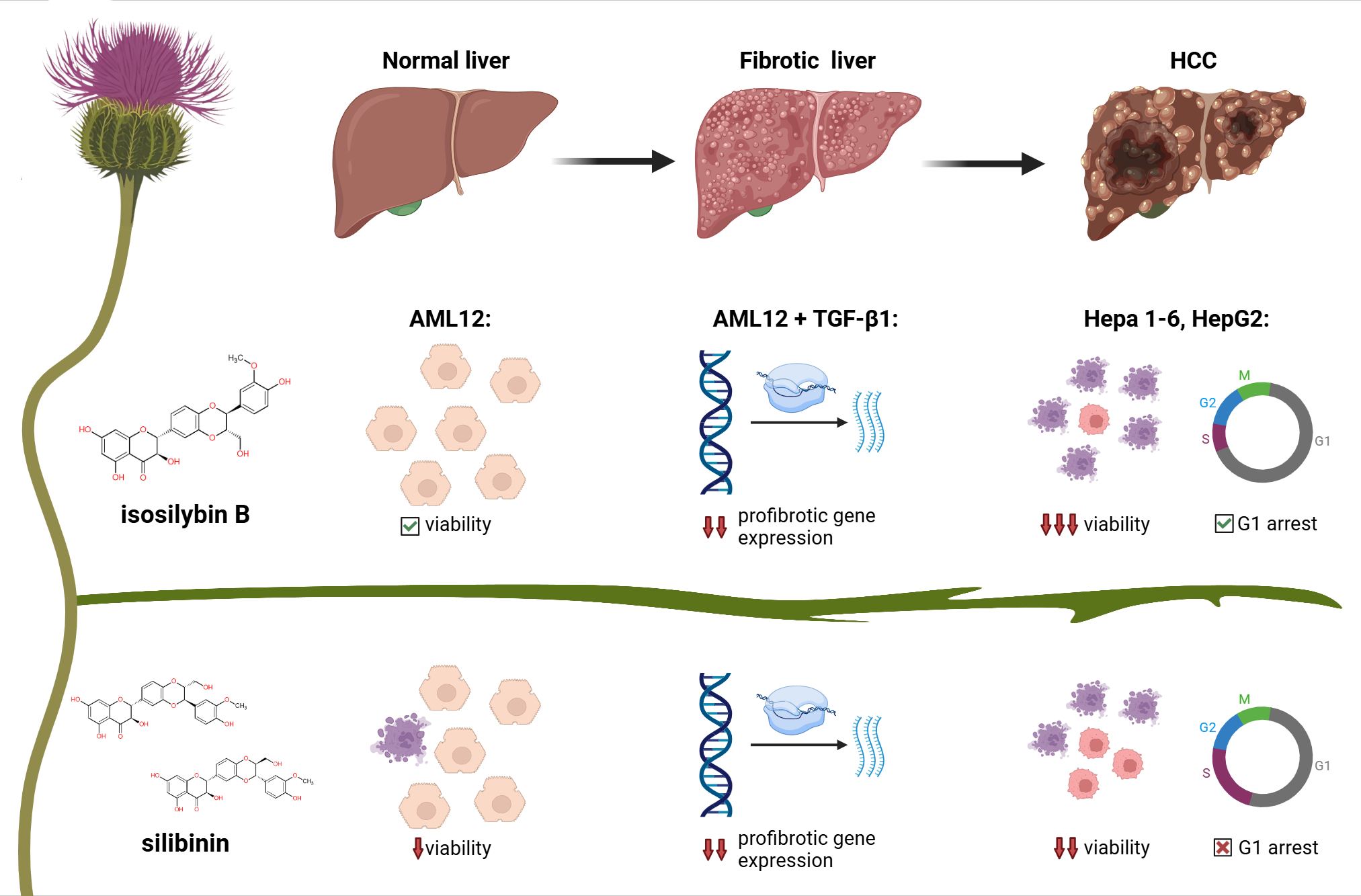
Figure 1: Isosilybin B from milk thistle shows stronger tumor selectivity and antifibrotic effects than silybin or whole silymarin. IB preserves normal hepatocyte viability, reduces profibrotic gene expression, and induces tumor-specific G1 arrest, suggesting advantages in the treatment of liver disease.
Follow the Topic
-
Discover Oncology

This is a fully open access general oncology journal that aims to provide a unified forum for researchers and clinicians. The journal spans from basic and translational science, to preclinical, clinical, and epidemiology, and welcomes content that interfaces at all levels of cancer research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Single-Cell RNA Sequencing in Cancer Immunotherapy
Cancer immunotherapy is a hot area of current oncology research, with its core focus on activating or enhancing the body's immune system's ability to recognize and kill cancer cells. However, cancer cells possess complex heterogeneity and dynamics, which affect the efficacy of immunotherapy in many ways. Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool in recent years, providing us with an unprecedented insight into the cellular heterogeneity and dynamics within tumors. This technology has revolutionized our understanding of cancer biology, especially in the context of cancer immunotherapy. By enabling researchers to analyze individual cells, scRNA-seq allows them to identify distinct cell populations, track cellular responses to treatments, and discover new therapeutic targets. This collection aims to compile cutting-edge research in this field and explore the various applications of single-cell RNA sequencing in cancer immunotherapy.
This collection will cover the following topics: 1. The latest advances in single-cell RNA sequencing technology in cancer immunotherapy, including research on technology optimization and data interpretation; 2. Using single-cell RNA sequencing to reveal the characteristics of immune cell subgroups in the tumor microenvironment and their interaction mechanisms with cancer cells; 3. Analyzing the molecular basis of immune therapy response and resistance through single-cell RNA sequencing, exploring new biomarkers and therapeutic targets; 4. Combining single-cell RNA sequencing with clinical studies of immunotherapy to assess treatment outcomes, predict patient prognosis, and optimize treatment plans.
Keywords: cancer immunotherapy, single-cell RNA sequencing, therapeutic targets, tumor microenvironment, treatment response
Publishing Model: Open Access
Deadline: Jun 30, 2026
Tumor Microenvironment
The immunological and stromal components of the tumor microenvironment (TME) play important roles in supporting or preventing tumor growth. The interaction of tumor cells and immune cells inside the TME is complex, and it can result in immune suppression, immune evasion, or, in the opposite case, successful immune-mediated tumor elimination. Cancer therapy has increasingly focused on targeting the TME. Immune checkpoint inhibitors (e.g., anti-PD1, anti-CTLA4), CART cell therapy, and vaccinations are all aimed at reactivating the immune system's ability to recognize and destroy tumor cells. Modulating immune component activity is being investigated as a way to counteract the immunosuppressive TME and boost immunotherapy efficacy. This thematic Collection will provide a thorough understanding of the complex role of the tumor microenvironment in the emergence of cancer treatment resistance, as well as innovative strategies for overcoming this challenge and investigating the tumor microenvironment's contribution to personalized medicine.
Keywords: tumor cells, TAMs, TILs, stromal cells, immunotherapy
Publishing Model: Open Access
Deadline: Jun 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in