
Siderophores are low molecular weight iron chelators produced by fungi to scavenge iron from the environment, and are essential for survival and virulence. The majority of siderophores produced by fungi belong to the hydroxamate class and are produced by non-ribosomal peptide synthetase (NRPS) enzymes, notorious for their highly unusual domain architectures. Work published in Nature Communications from our laboratories uncovers the cryptic programming events the NRPS enzyme responsible for production of the ferricrocin siderophore. This work highlights new capabilities of NRPS enzymes allowing biosynthetic assignment of other cryptic NPRSs and setting the stage for bioengineering efforts.
This collaborative effort was fantastic, but it nearly didn’t happen at all. Rewind to March 2018, and Prof. Yi Tang was visiting the University of Warwick to deliver the annual Warwick Chemical Biology Lecture. It was snowing (one form of weather the UK is particularly ill-prepared for), and I was stuck in traffic trying desperately to make it in time for my scheduled meeting with Yi that morning. I didn’t make it. Luckily, we managed to find time in the afternoon to talk science, where we discussed some recent work applying intact protein mass spectrometry to polyketide synthases. Thinking this approach might be helpful, Yi put me in touch with Dr. Yang Hai who was a LSRF Fellow in his lab at UCLA at the time. Several e-mails and Zoom calls later, Yang and I are working on 2 different projects related to fungal siderophore biosynthesis. Sending samples across the Atlantic, we managed to make some quick progress on the SidD NRPS, responsible for assembly of the fusarinine siderophore (Chem. Sci., 2020, 11, 11525). However, the second project focussed on understanding the biosynthesis of ferricrocin by the SidC NRPS - this proved a more challenging prospect!
In bacteria, NRPS enzymes tend to exhibit a colinear relationship between the order of enzymatic domains and the final product. This, combined with the ability to predict the individual amino acids from the primary sequence of each adenylation (A) domain, allows prediction of peptide structure from sequence data alone. The tyrocidine NRPS is a good example of a ‘textbook’ NRPS system (Fig. 1a). In contrast, fungal NRPSs exhibit unusual characteristics that make understanding their biosynthetic logic more challenging. Firstly, prediction of A domain specificity from sequence data is not possible in fungi – suggesting different evolutionary origins. In addition, modules lacking dedicated A domains are commonplace, implying that the thiolation (T) domain must be loaded by an A domain from a different module. Further complicating matters, the sequence of the peptide product often doesn’t correlate to the order of domains, suggesting nonlinear behaviour. The SidC NRPS is a perfect exemplification of all these unusual features (Fig. 1b).
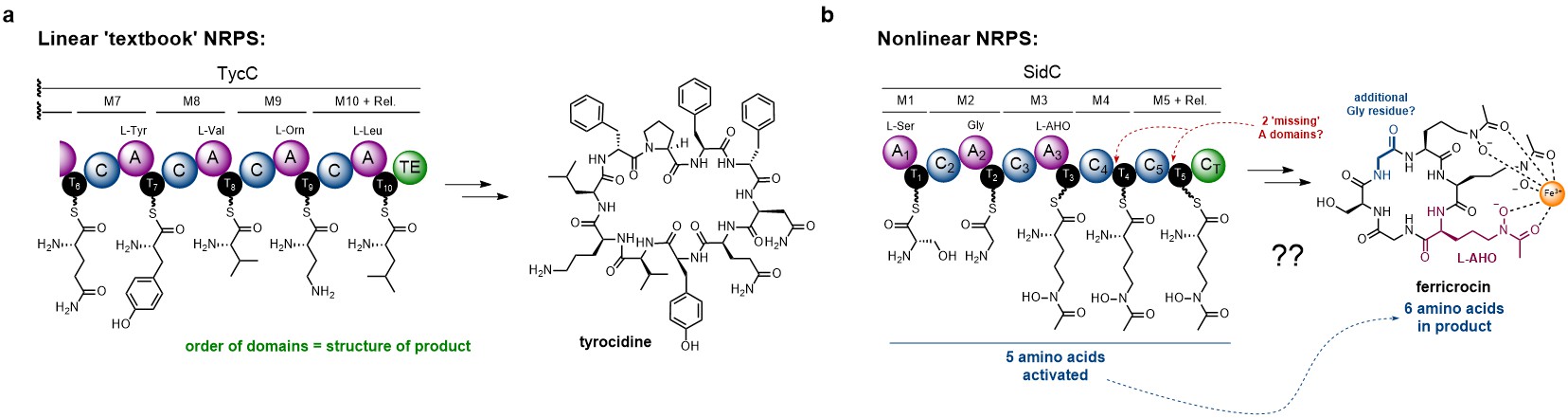
Figure 1. Examples of linear vs nonlinear NRPSs. a). Partial domain and module organisation of the NRPS responsible for production of tyrocidine, showing the individual amino acids loaded onto each T domain. b). Domain organisation of the SidC NRPS responsible for the biosynthesis of ferricrocin. The non-canonical features are annotated in red and blue. Domain abbreviations are as follows: A = adenylation; C = condensation; T = thiolation.
Two features of the SidC NRPS were of interest – i). the lack of A domains in modules 4 and 5; and ii). a missing set of domains for incorporation of the second Gly unit (Fig. 1a, highlighted in blue). Going into this project, Yang had managed to overproduce and purify the entire SidC NRPS from yeast (520 kDa), allowing some rigorous biochemical characterisation. Collaborating with researchers in the California area (Garg Lab – chemical synthesis; Zhang / Loo Labs – A domain specificity / loading assays), the recombinant enzyme was shown to produce ferricrocin in vitro, and the specificity of the A domains could be assigned (Fig. 1b). However, this still left the mechanisms behind the usual features of the SidC NRPS unanswered – and the start of our work together.
By virtue of the biosynthetic intermediates being covalently tethered to the enzyme (via a 4’-phosphopantetheine arm), changes to substrates bound to the T domain can be monitored by intact protein mass spectrometry (MS). Using the right instrumentation, mass shifts associated with gain / loss of biosynthetic intermediates can be readily tracked on large, multidomain constructs – an approach we regularly use in the Jenner Lab. Application of this approach allowed us to unambiguously prove that the A3 domain was capable of loading the hydroxamate-containing Nδ-acetyl-Nδ-hydroxy-L-ornithine (L-AHO) amino acid on the T4 and T5 domains, in an inter-modular manner. It appeared that the corresponding N-terminal condensation (C) domain was critical for these loading events, and an AlphaFold model of this region suggested that the C domains form an interface with the A3 domain, bringing the T domains into close proximity (Fig. 2a).
Interestingly, first clues to the origin of the second Gly residue had been sitting right under our noses for some time. One of the first assays we conducted was to monitor Gly loading on a C2A2T2 fragment. Expecting to see a single +57 Da peak corresponding to one Gly unit, we instead saw a mixture of peaks which appeared to correlate to tri-Gly and penta-Gly polymeric chains. Unsure what this meant at the time, we focussed our efforts elsewhere on the NRPS. It was Yang who dug out the data sometime later and suggested that the module – or more specifically the A2 domain – might be catalysing amide bond formation between free α-NH2 group and a Gly residue (Fig. 2b). This was reminiscent of activity observed for a stand-alone A domain in streptothricin biosynthesis, which instead uses the free ε-NH2 of a β-lysine residue (Nat. Chem. Biol., 2012, 8, 791). Some careful time-resolved experiments allowed us to monitor this process, and also show that the downstream C3 domain acts as a gatekeeper for this process – only accepting a three-residue chain (Fig. 2b).
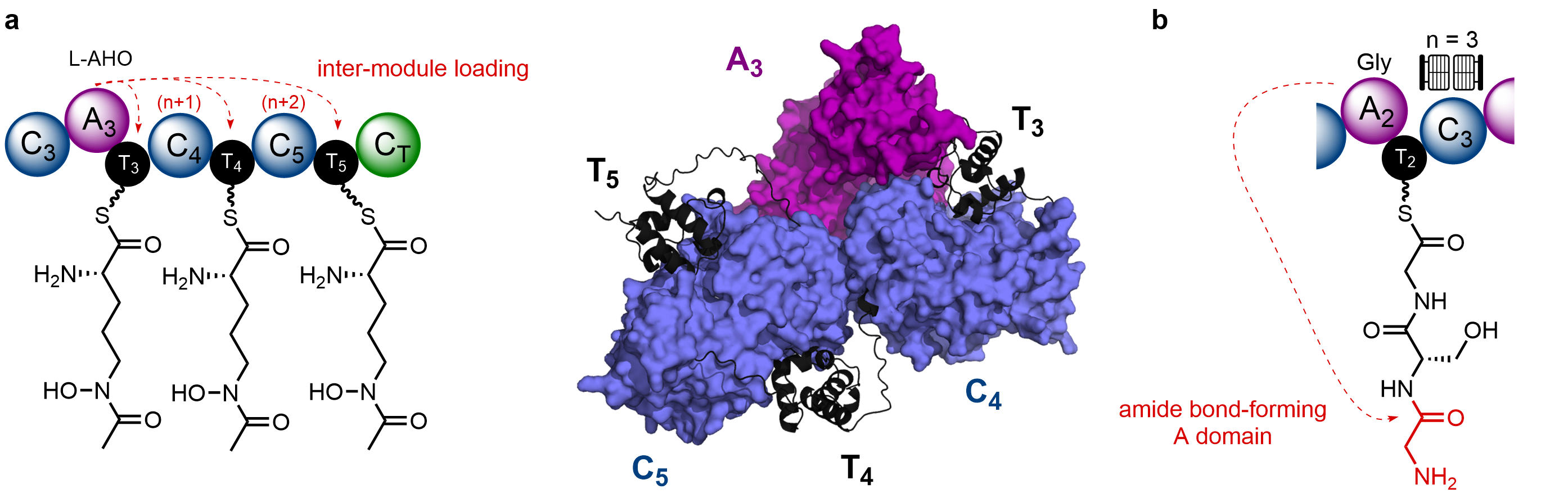
Figure 2. Cryptic programming events in the SidC NRPS. a). The A3 domain loads L-AHO to the T3 domain (intra-modular), in addition to the T4 and T5 domains (inter-modular). An AlphaFold model of the A3T3C4T4C5T5 region suggests close packing between the A3 domain and C4/C5 domains allows substrate transfer to the T domains. b). The A2 domain catalyses amide bond formation with the terminal NH2 group, to form a three-residue chain. The downstream C3 domain acts as a gatekeeper for this process.
This work was full of surprises and highlights some of Nature’s ingenious approaches to enzyme efficiency. It also sets the stage for some exciting work exploiting this NRPS, which we are actively looking at. On a personal level, this was some of the most enjoyable science I have ever done - there was something quite exhilarating about Yang and I tackling this puzzle together, yet never having met each other in person. However, just before the pandemic struck, I managed to visit Yang and Yi in LA, where highlights included rooftop drinking, a visit to Griffith observatory and some questionable karaoke (ask Yi for the video evidence…).

Very fond memories with friends and collaborators, but as I said at the start; it almost didn’t happen at all.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in