Looking for clues in cyanobacterial death
Published in Microbiology

Cyanobacteria are a group of bacteria that can perform photosynthesis, requiring just light and carbon dioxide to grow. They are found globally, frequently in freshwater and marine environments, and often form dense multi-cellular communities like mats or blooms. These dense growths commonly undergo massive die-offs where the cyanobacterial cells disintegrate rapidly, releasing organic compounds into their environment and fueling rapid consumption by heterotrophic organisms. This results in extreme oxygen drawdown, effectively suffocating the surrounding aquatic life. Occasionally, toxins can also be released by cyanobacteria, creating further concerns for human and animal health. Despite the consequences of these mass die-offs, the cause of this large-scale cyanobacterial death is largely unknown.
Our study aimed to address this unknown by focusing on a cyanobacterial species that readily grows in dense mats, Candidatus Phormidium alkaliphilum. In our laboratory, we have been growing an enrichment culture, containing multiple bacterial species but with Ca. P. alkaliphilum as the dominant member, sourced from microbial mats from a highly alkaline Canadian soda lake. We had noticed that when left in a dark place for a few days (e.g., a cupboard), samples of this primarily green culture would turn an intense blue colour, suggesting that the internal blue pigment, phycocyanin, had been released from the cyanobacterial cells. Further observations proved that this process could be consistently repeated, and microscopy showed that the cyanobacterial cells had indeed lysed.
Although we could observe that the cells were lysing, the mechanism for this phenomenon remained elusive. We decided to systematically investigate this process by incubating the culture in dark and anoxic conditions for 12 days. We took samples every two days from the culture biomass (solids) and the liquid media (supernatant). For each sample, we measured phycocyanin concentration, and performed metagenomics and metaproteomics to observe the microbial community dynamics at the level of DNA and protein respectively. We had three main hypotheses for cause of death: 1) the cyanobacterial cells died from predation or antagonism by neighbouring bacteria or protists, 2) the cyanobacterial cells died from viral infection, and 3) the cyanobacterial cells died from a programmed cell death response.
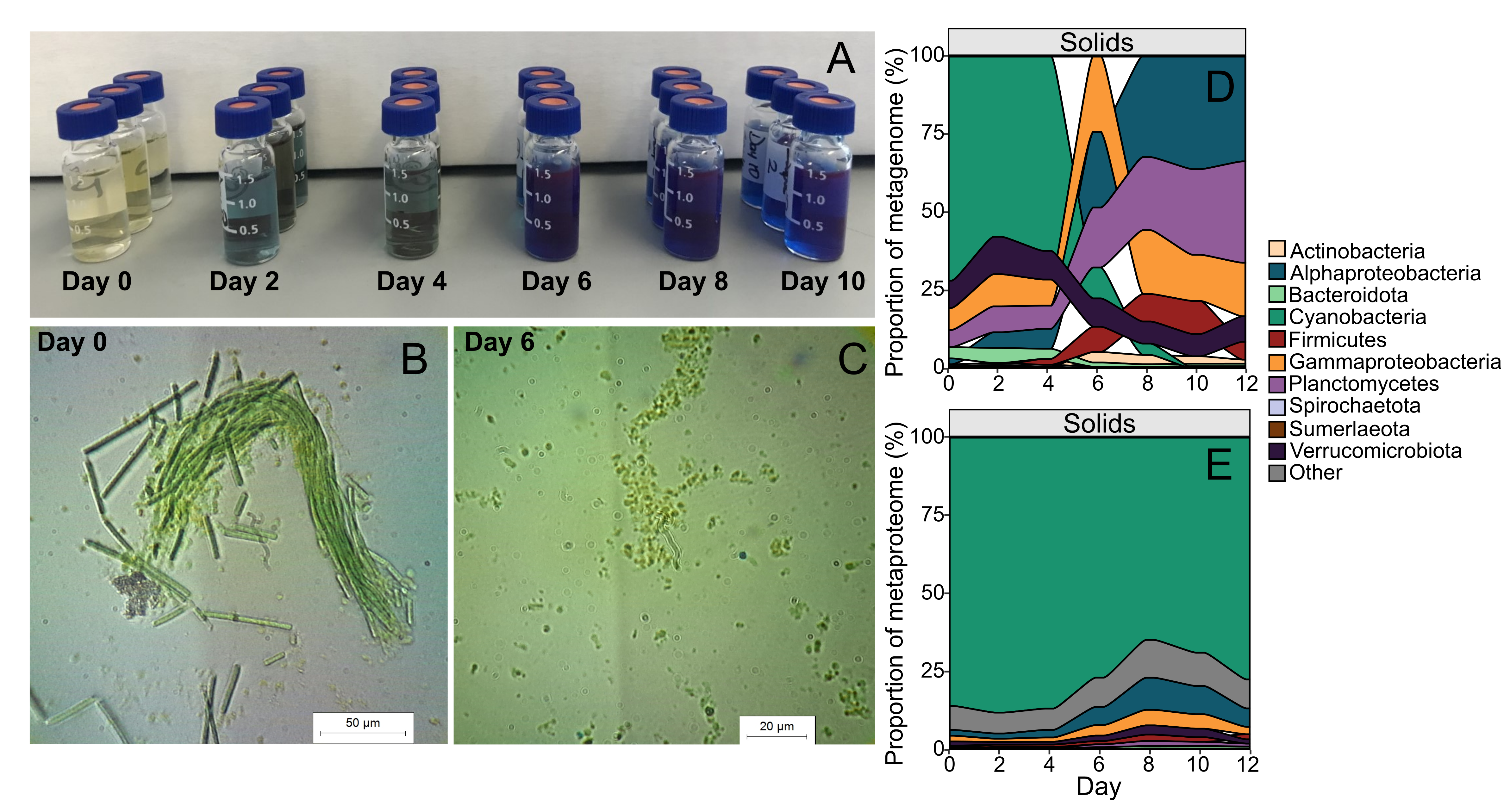
We observed through phycocyanin concentration in the media, that the majority of cell lysis occurred around day 6 (Figure 1A-C). This lysis coincided with a dramatic decrease in total cyanobacterial DNA (Figure 1D), but hardly any change in total cyanobacterial proteins (Figure 1E), suggesting that cyanobacterial DNA was rapidly degraded, but cyanobacterial proteins remained comparatively untouched. From the metagenomics data, we did not observe any increase in protist or viral DNA in proportion to the cyanobacterial demise (Figure 2). We also did not observe a corresponding proportional increase in protein or DNA content of any of the heterotrophic bacteria in the community (Figure 1DE). This data suggested that other bacteria, protists, and viruses were not likely contributors to the cyanobacterial death, and so hypotheses 1 and 2 could be largely dismissed.
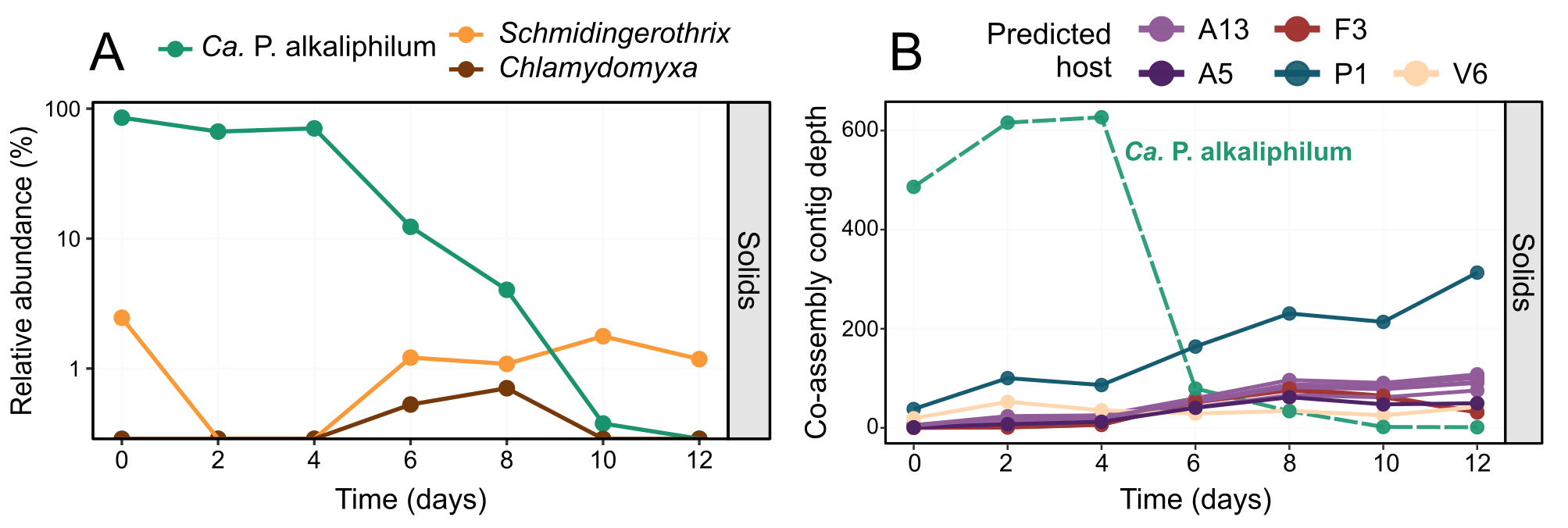
Figure 2. Dynamics of protist and viral sequences during incubation. Relative abundance of Ca. P. alkaliphilum (green) and the two protist species identified (orange and brown), over the course of the incubation (A). Abundance of viral associated contigs over the incubation coloured by their potential bacterial host (B). A13: Rhodobacteraceae, A5: Pararhodobacter, F3: Alkalibacterium, P1: UBA6054 (genus of Planctomycetota), V6: Verruco-01 (family of Verrucomicrobiota). The green dashed line shows the average contig depth for the Ca. P. alkaliphilum co-assembly MAG over the same samples.
We then turned our attention to hypothesis 3, a programmed response leading to cell death in the cyanobacteria. From our data, the rapid disappearance of cyanobacterial DNA already supported this hypothesis, as DNA fragmentation is a commonly observed phenomenon in cells undergoing programmed cell death. The cyanobacterial proteome data held further clues and we noticed concerted changes in certain functional categories of proteins. Proteins related to transcription and translation decreased greatly in abundance prior to cell death (Figure 3A). In cells that had just lysed (post-lysis supernatant fraction) we observed an increase in proteases and proteins involved in amino acid metabolism, suggesting active remodelling of the cyanobacteria proteome (Figure 3D). We also observed multiple toxin-antitoxin systems, including a putative system in which antitoxin expression decreased before lysis, and the putative toxin expression increased post-lysis (Figure 3B). An operon belonging to the effector complex of a Type III CRISPR-Cas system was also upregulated post-lysis (Figure 3C). Since no evidence of viral activity was found, it’s possible that this CRISPR-Cas system might be operating in the programmed cell death response.
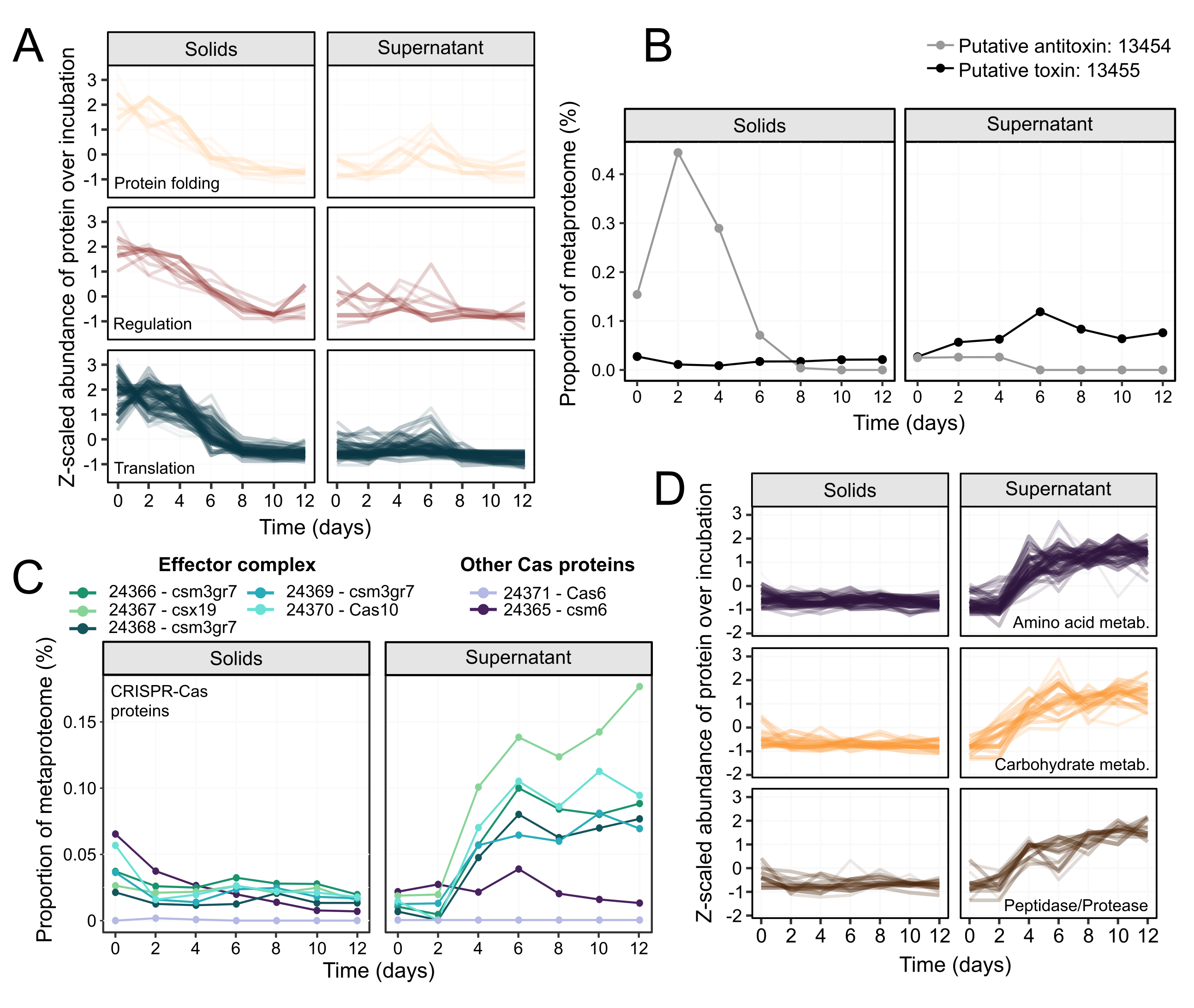
Figure 3. Dynamics of Ca. P. alkaliphilum proteins over the course of the incubation. Proteins from functional categories that were significantly more abundant in the solids fraction before cell lysis (A). Metaproteome abundance of the prospective toxin-antitoxin protein pair over the incubation (B). Abundance of proteins located in a CRISPR-Cas operon that were significantly enriched in the supernatant post-lysis (C). Proteins from functional categories that were significantly higher in abundance in the post-lysis supernatant fraction (D).
Based on these observations, we propose that the most likely cause of the mass cyanobacterial die-off in our system was programmed cell death in response to severe energy limitation brought about by darkness and anoxia. Our study gives unique insight into the molecular response of a bacterial cell undergoing cell death and has provided several potential death-mediating protein candidates for further research. Additionally, the encoded nature of this cell death response raises questions as to the ecological role of this process, and the evolutionary advantage that the cyanobacteria gain by preserving this pathway in nature. Based on existing literature, it is possible that the majority of cyanobacterial cells die, while a few “persister” cells remain in a dormant state. Released compounds from the lysed cells could further supplement survival of the persister cells until conditions improve and growth can resume. In their natural setting, these cyanobacteria frequently experience periods of prolonged darkness and anoxia due to sinking within microbial mats or sediments, and therefore the ability to withstand periods of energy starvation is an important survival strategy. Future work will be important to investigate if this mechanism extends to other densely growing cyanobacterial species, and if it could be harnessed to control harmful blooms or to aid bioproduct production.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in