Microbial sentries of chemical balance in the rhizosphere
Published in Microbiology

Plants maintain a microbiota – a diverse collection of heterotrophic microorganisms which are intimately associated with it, and depend on carbon that it fixes. Many of these microbes have evolved to manipulate the plant into providing a more habitable environment for themselves. Some of these interactions are mutualistic, and the microbes reciprocate by providing some benefit to the plant, but many plant-microbe interactions are costly for the plant and can compromise plant productivity. This is most apparent in cases of acute plant disease, but we expect that plants endure many “chronic infections” that carry more subtle costs that are not easily diagnosed. To complicate things further, microbial effects on the plant are not always direct. Every member of the microbiota interacts not only with the plant, but also with other microbes, and these interactions can result in emergent properties with either positive or negative consequences for the host.
How do we tease apart the mutualists in the community from the parasites? And how do we measure the net effect of all interactions within the microbiota on the plant?
To address these questions, we applied what we call “top down community deconstruction”. We began our work with a gnotobiotic microcosm containing a host plant and a 185-member bacterial synthetic community which serves as a model for a natural plant microbiota. We first studied this community as a whole, understanding how it responds to varying abiotic conditions. We then used this information to deconstruct the community into defined modules – groups of co-occurring bacterial strains, and inoculated the plant with each of these modules separately – to see how each module affects the plant – or in combination with other modules – to examine how do the different modules interact with each other. We found that some modules induced very strong effects in the plant, which were clearly manifested by root growth inhibition (RGI). We were particularly captivated by the observation that other modules were able to completely suppress this effect, such that when the full community is present root growth is not inhibited. This allowed us to conclude that plant-microbiota interactions are indeed non-additive, but rather governed by epistasis.
At this point, we postulated that we could find a ‘master regulator’ taxon. One whose presence in the community is necessary and sufficient to suppress RGI and enforce stereotypic root development. Finding this taxon required some detective work using a reduced tripartite system with only two microbial strains at a time. This detective work pointed to a clear candidate – strains from the genus Variovorax appeared to be able to revert RGI induced by a diversity of other strains.
The ultimate test, however, for the role Variovorax play in the community, is a drop-out experiment – comparing the effect of the community on the plant with or without Variovorax present. And indeed, delighting us once more, dropping Variovorax out of the community (or rather, not including them in the inoculum), caused RGI, proving that Variovorax are necessary and sufficient for enforcing stereotypic root development. This observation is not idiosyncratic. Variovorax revert RGI under multiple substrates, biotic and abiotic context and appear to be a core taxon in the plant microbiome across species and geography.
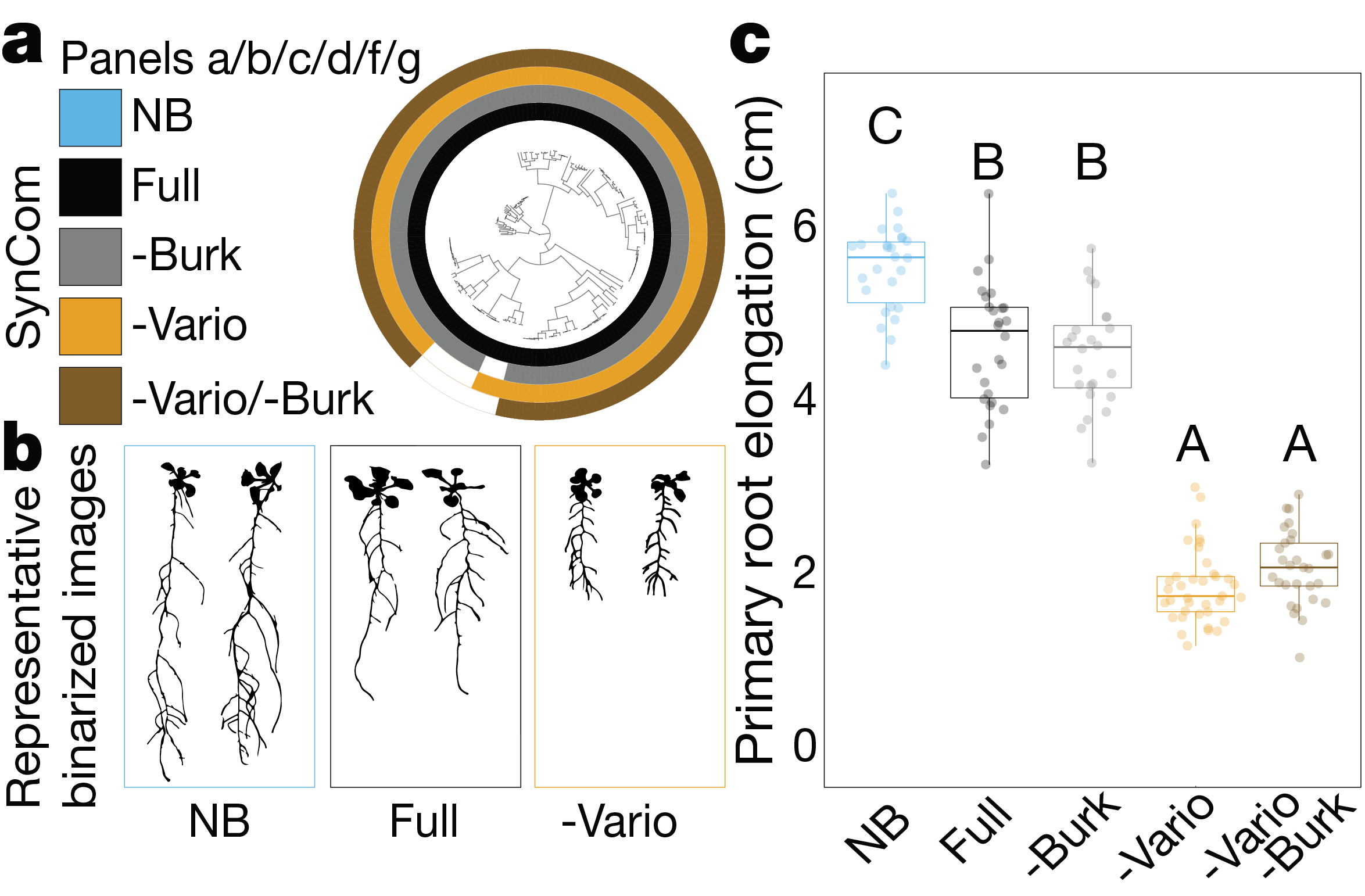
This discovery unlocked a new research path for us, transforming our study from the ecological to the mechanistic realm. We soon realized that Variovorax can revert RGI induced by the phytohormones auxin and ethylene and that auxin-related gene expression was induced by RGI-inducing bacteria, and reduced when Variovorax were present. This led us to hypothesize that while multiple bacterial species hijack plant development by flooding it with auxin, Variovorax mitigate this effect by effectively metabolizing this excess auxin. We refer to this as ‘metabolic signal interference’. Comparative genomics helped us identify loci in the Variovorax genome that are involved in this process. Through functional screening of the Variovorax genome, validated by both gain- and loss-of function assays in our bacterial strains, we were able to pinpoint the exact locus in the Variovorax genome that enables this phenotype and are now working to characterize this auxin degradation operon.
This study reinforces the dominance of auxin in plant-microbe communication and demonstrates that it is now feasible to mechanistically study host-microbiota as a whole, rather than as a discreet list of host-microbe interactions.
This project was a group effort within the Dangl lab and it was made possible through significant contributions by all of the paper's authors. We are greatful to them all.
Cover photo credit: Dr. Connor R. Fitzpatrick
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in