
Multidrug resistance in Neisseria gonorrhoeae, the bacterial pathogen responsible for causing gonorrhea, is rapidly increasing. Our understanding of the genetic basis for resistance is incomplete for azithromycin, which is a commonly used macrolide antibiotic that targets the bacterial ribosome and was the focus for this study.
To discover new azithromycin resistance mutations, we conducted a genome-wide association study (i.e., comparison of susceptible and resistant genomes in order to find distinguishing mutations) on a dataset of ~4500 strains. We picked up mutations in the 23S rRNA, a component of the ribosome; this was a good positive control because 23S rRNA mutations are prevalent, known contributors to azithromycin resistance in N. gonorrhoeae.
Looking down the list at other significant genes, we saw many that had not been previously associated with macrolide resistance in any bacteria, which was a troubling sign. In general, it’s not too hard to run a GWAS or a machine learning algorithm and come up with a lot of “novel” resistance genes, and it’s important to think through your model and the underlying biology to figure out whether they’re causal or – more likely – statistical artifacts. It turns out that most of these significant genes were genetically linked to the 23S rRNA mutations (i.e., strains that had these mutations also had 23S rRNA mutations), which led us to think that these associations were false positives. Additionally, we didn’t pick up other known resistance mutations in efflux pumps that had been previously characterized. In other words, both the precision (among the significant GWAS hits, what fraction were true positives?) and the recall (how many of the true positives do we actually pick up?) were low in the initial GWAS, providing limited confidence in our results.
To address this, we repeated GWAS controlling for the presence of these 23S rRNA mutations. This is equivalent to asking: after accounting for the contributions of the 23S rRNA mutations to azithromycin resistance, what variants are now significant? The idea of conditioning on known effects has been commonly used in the human GWAS field, but is not as widely used for bacterial GWAS. Conditional GWAS was very effective for us: we saw that the probable false positives disappeared, and known true positives jumped up in significance (Figure 1). The bottom right quadrant are variants significant in the first GWAS that were no longer significant in the conditional GWAS, whereas the top left quadrant is the opposite: variants that weren’t significant in the first GWAS but became significant after conditioning.
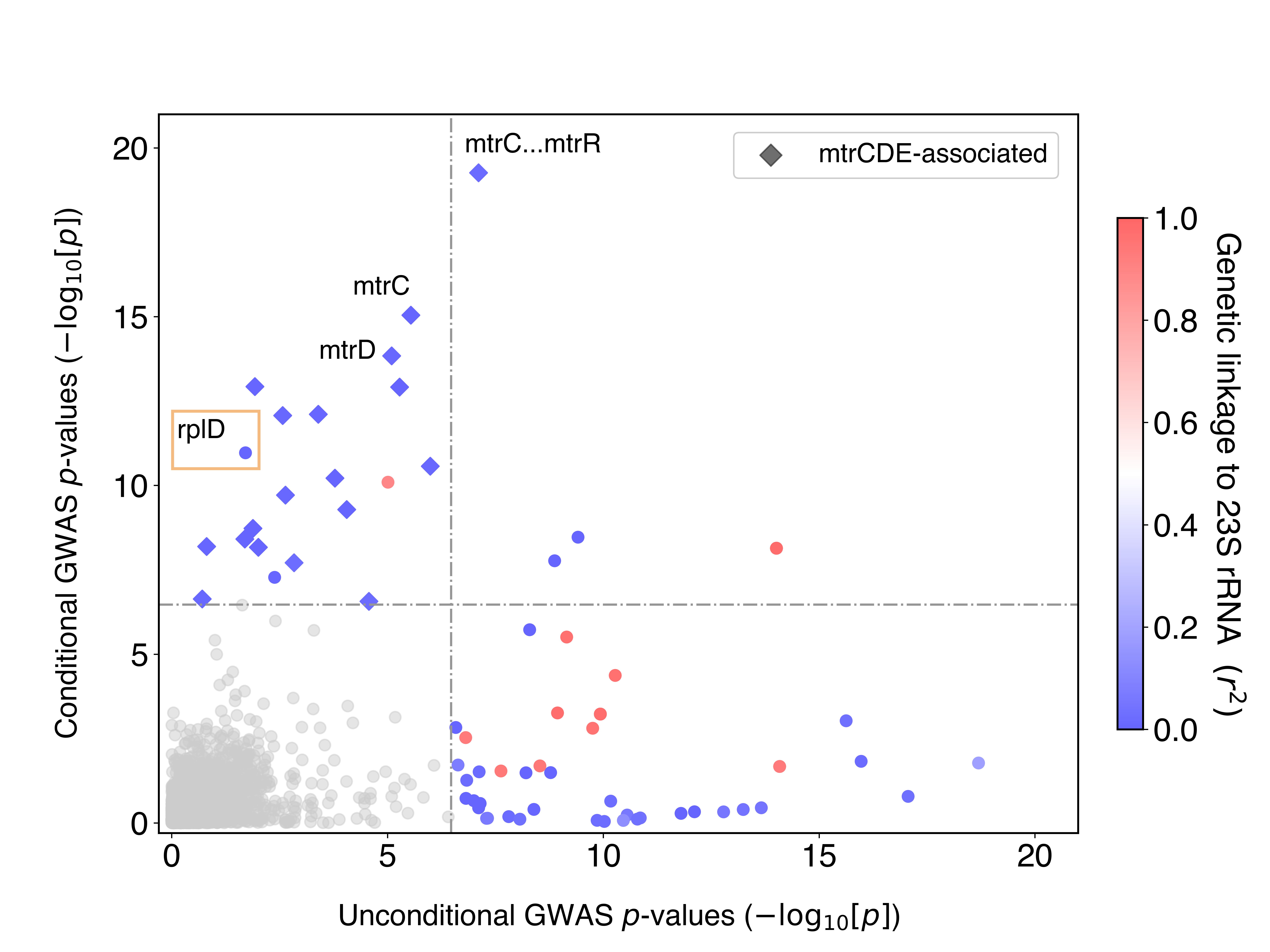
Figure 1 – GWAS conditional on 23S rRNA mutations and dataset demonstrates decreased confounding and increased power.
The increase in precision and recall of the conditional GWAS approach gave us more confidence in the other significant genes. We followed up by looking at the most significant variant after the known resistance mutations, which mapped to a G70D mutation in RplD, an important 50S ribosomal protein that azithromycin is known to bind to. The wild-type amino acid, glycine, is non-polar with a hydrogen tail and becomes mutated to the negatively charged amino acid aspartic acid in resistant strains. We think this mutation sterically hinders macrolide binding and allows ribosomes in N. gonorrhoeae to continue functioning (Figure 2).
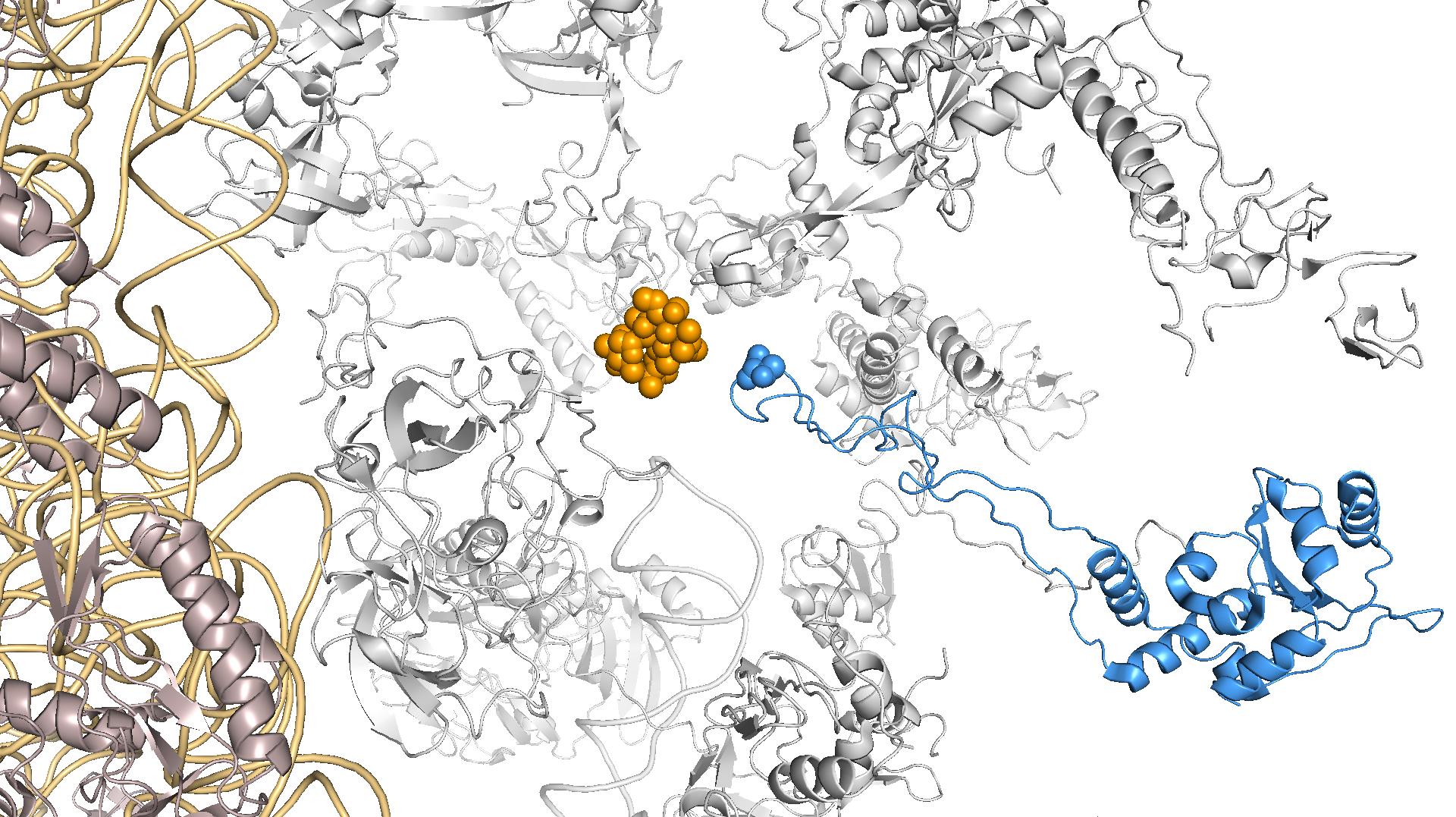
Figure 2 – RplD G70 is part of the azithromycin binding pocket in the 50S ribosome (structure used is from Thermus thermophilus).
Homologous mutations increase resistance in other bacteria and the G70D mutation has been previously observed in N. gonorrhoeae, but confusingly, previous studies weren’t able to show a clear association with resistance. To confirm our GWAS results, we made isogenic N. gonorrhoeae mutants of RplD G70D in the lab and showed that they indeed increased macrolide resistance. We also measured fitness cost by comparing growth of the wildtype N. gonorrhoeae strain and the G70D transformant in rich media, but observed no significant growth differences between the strains. Since other macrolide resistance mutations are typically associated with fitness costs, a follow up experiment would be to repeat this assay in other environmental conditions.
Finally, we took a targeted look to see if there were other RplD mutations associated with resistance that would be too rare to find in the GWAS. We found numerous novel mutations, all of which appeared associated with even higher macrolide resistance than the G70D (Figure 3). The occurrence of repeated mutations in one particular spot in a protein often suggests that there’s strong selection at that site, and gave us increased confidence that selection due to antibiotic pressures was indeed acting on RplD. We think those other mutations will also be important for drug resistance surveillance efforts in the future as azithromycin use continues to increase. It would also be interesting to look at the fitness cost of the other novel RplD mutations; we hypothesize that these novel mutations may have higher fitness costs than the G70D mutation since they are seldom observed in the population.
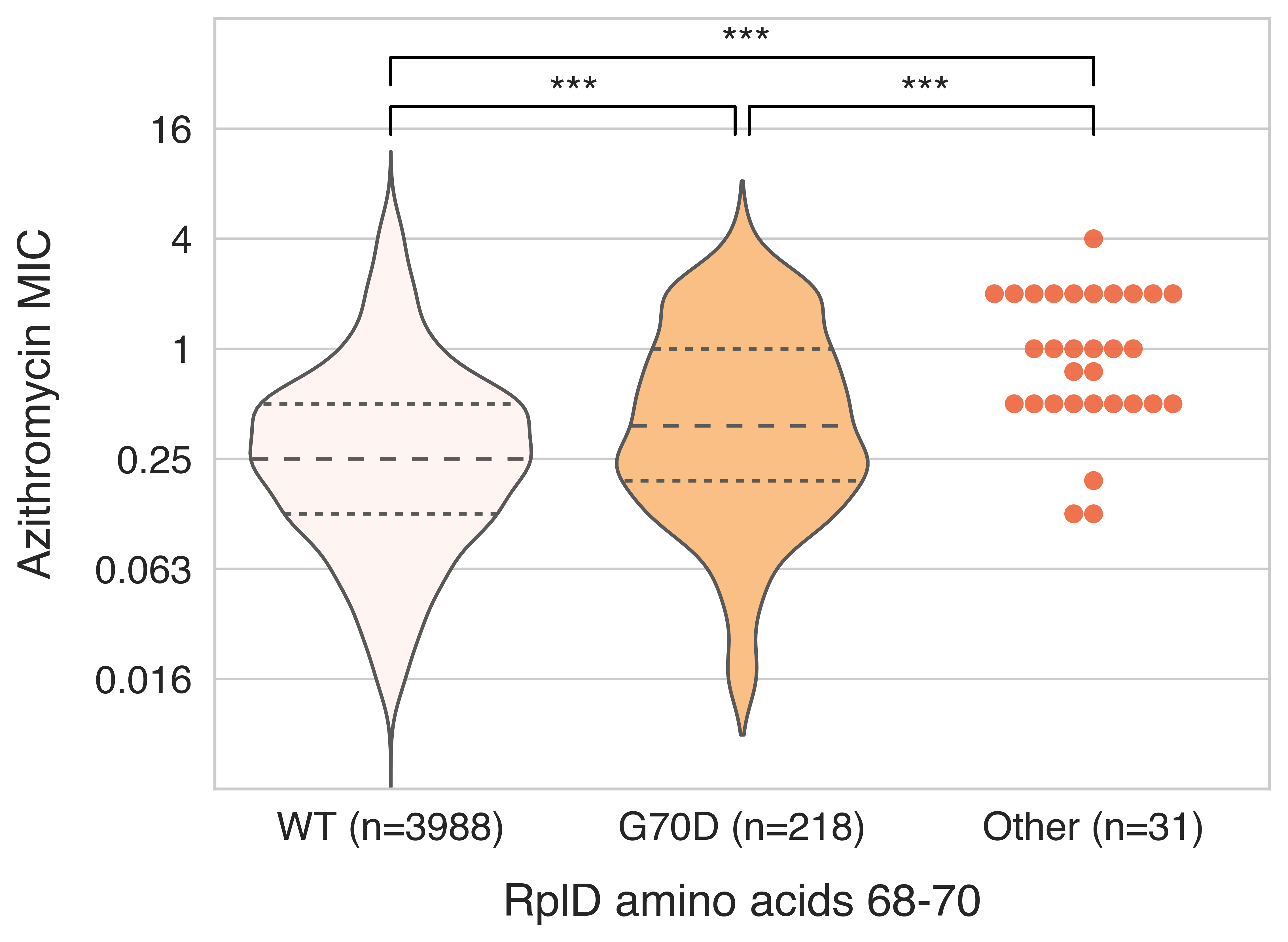
Figure 3 – Distribution of azithromycin MICs for RplD macrolide binding site variants.
The full paper (“Increased power from conditional bacterial genome-wide association identifies macrolide resistance mutations in Neisseria gonorrhoeae”) is available at Nature Communications: https://www.nature.com/articles/s41467-020-19250-6
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in