Playing "Hide and Seek" with transferred mRNA and nanotubes
Published in Protocols & Methods
This is the story of my MiMB paper: "Detection of mRNA Transfer Between Mammalian Cells in Coculture by Single-Molecule Fluorescent In Situ Hybridization (smFISH)".
In 2010 I began to look for a postdoc position. I met Rob Singer at an EMBO RNA meeting in Israel and we had an interesting discussion on the possibility of RNA transfer between cells. I was inspired by recent research on RNA in exosomes in mammals, and mRNA transport through plasmodesmata in plants. Rob is the father of single-molecule RNA imaging: he developed both single-molecule FISH and live imaging of RNA molecules using MS2, and many more cool tools for RNA imaging. My idea was to use these tools to follow mRNA from cell to cell. I chose to concentrate on mRNA, because by that time there was an explosion of research on extracellular miRNA, but little work done on extracellular mRNA.
What I found, eventually, was that mRNA can transfer between mammalian cells through a unique cell-to-cell conduit called membrane nanotubes (a.k.a tunneling nanotubes, TNTs), and not by diffusion. After five years of work, which included a move back to Israel where I joined Jeff Gerst’s lab, and a collaboration with Arjun Raj, we finally published our work in PNAS. I wrote a post “Behind the Scenes” in my personal blog describing the work and the hardships of getting it done.
Briefly, I used model cells – mouse embryonic fibroblasts (MEFs) – in which the endogenous β-actin mRNA was labeled with 24 repeats of the MS2 stem-loop aptamer (MS2 coat protein binding sequence – MBS; I call the cells MBS-MEFs). These donor cells were co-cultured with wild-type (WT) MEFs which acted as acceptor cells. By using FISH with MBS-specific probes, I could detect transfer of the β-actin-MBS mRNA. We also showed transfer of endogenous untagged mRNAs in other co-culture systems.
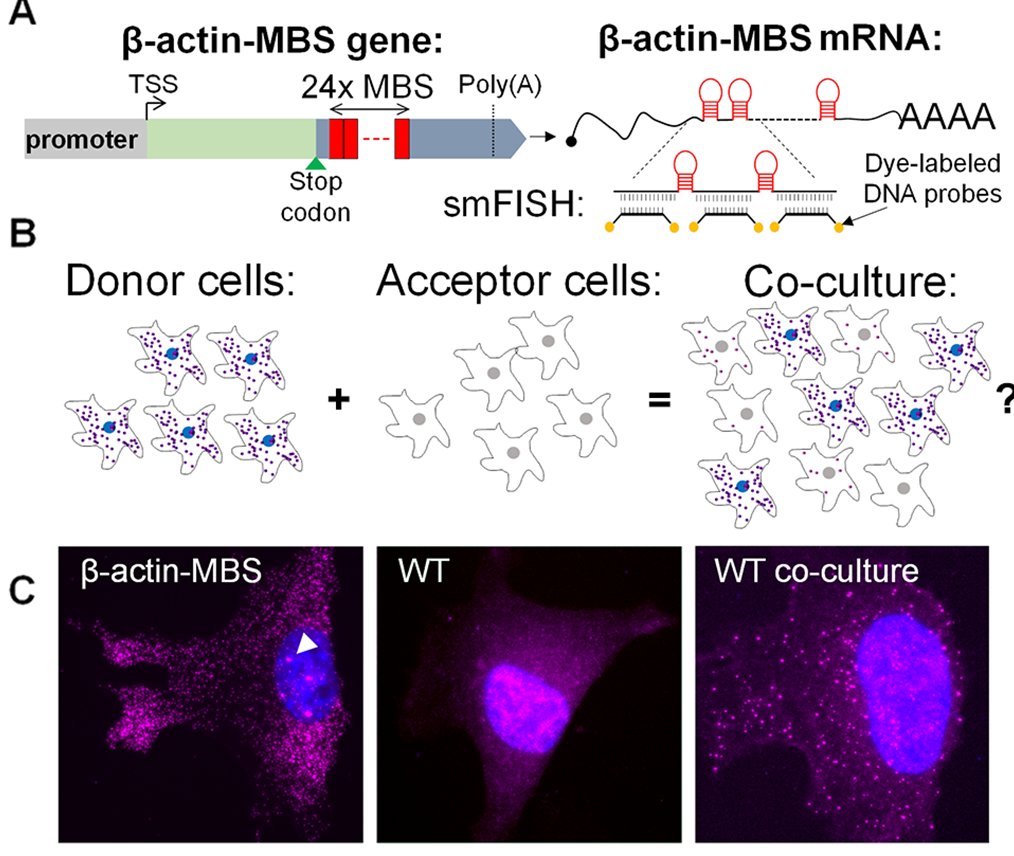
One of the reasons that publication took so long is because the MS2 system failed me. The MS2 system is over 20 years old, but still the best method to follow single mRNA molecules in live cells. It is a two-part system – the MBS in the mRNA, and the MS2 coat protein fused to a fluorescent protein (MCP-GFP). If the mRNA has multiple repeats of the MS2 aptamer, it will bind multiple MCP-GFPs, and the mRNA is seen as a discrete spot moving about the cell. I dreamt of following mRNA molecules in live cells, moving from cell to cell, so I could get information about the rate of transfer, the speed at which these mRNAs move, maybe test transfer of the mRNA with other cellular components (proteins, organelles).
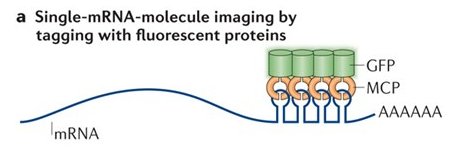
As it turns out, expressing the MCP-GFP in the MBS-MEFs inhibited the transfer of β-actin-MBS mRNA. I tried other live imaging systems, such as Rcas9, Riboglow or FIT-probes – but these did not work well in my hands.
I decided to forget about live imaging and go back to FISH – which works very well in detecting the transferred mRNA. The idea was to do FISH combined with immunofluorescence to detect mRNA inside nanotubes and see if it is co-transported with certain proteins or vesicles. However, here is the problem – the FISH protocol is rather destructive for the delicate nanotubes. Nanotubes are very thin, and on top of that – they do not attach to the surface of the plate like filopodia. Imagine thin threads connecting cells hanging in the air (well, liquid) without support. It is relatively rare to preserve them in the regular FISH protocol.
The game of Hide and Seek started when I started to modify the FISH protocol. I wanted to make protocol less destructive or to make the nanotubes more stable and resilient.
The first thing I tried was to reduce the number of washes, and to use a pipet aid at low speed, instead of vacuum aspirator when I changed buffers. That made very little difference. I tried a protocol for IF from Emil Lou’s lab who study nanotubes, but that also had little effect.
I decided to use a more stringent fixation to strengthen the nanotubes. Adding 0.5% of glutaraldehyde during fixation (which is typically performed with 4% paraformaldehyde) did the trick! I managed to increase ~4-fold the number of detected nanotubes.
Approximately at that point in time, Yaron Shav-tal asked me if I wish to write a method paper for MiMB on detection of mRNA transfer through nanotubes. I was so excited from the success, that I thought it required just a little more optimization, so sure I will write this method paper. I had 8 months to the deadline – plenty of time. Little did I know that the game has just started.
I did a co-culture experiment followed by FISH with glutaraldehyde to detect the mRNA transfer and preserve the nanotubes. I got no mRNA transfer. Zero. That didn’t make any sense. But ok, the glutaraldehyde increases the autofluorescence and that might interfere with the FISH spot detection. I tried several approaches to reduce the authofluorescence: playing with glutaraldehyde concentration, switching the FISH probe to a far-red dye where there’s low autofluorecence, and adding different oxidants. Eventually, sodium borohydride (NaBH4) did the trick and completely alleviated the autofluorescence. NaBH4 also fizzles like carbonated water. This did not go well with the nanotubes, but it was still better than the original FISH protocol. Except I still did not detect any transferred mRNA. Obviously, glutaraldehyde did something bad to the FISH.
I looked for alternative buffers that are supposed to preserve the cytoskeleton, maintain filopodia, but use paraformaldehyde (like in FISH). This time, the FISH worked well. I could detect transferred mRNA as well as with the regular FISH. But guess what? Nanotubes count was down again to the original level.
At this point, it was obvious that something is hiding the transferred mRNA from being detected (but not endogenous mRNA, or the mRNA in the donor) when I fix the cells with glutaraldehyde.
I started to use brute force to get rid of that (most likely protein) obstacle.
I took several approaches that are aimed at unfolding or digesting proteins.
The deadline came and went and Yaron asked me to write something, so I wrote a less innovative protocol for detection of transferred mRNA by FISH. Nevertheless, I kept trying, and finally hit a breakthrough. Using either proteinase K to pre-digest the samples prior to hybridization, or using 1M urea instead of 10% formamide during hybridization resulted in a small but detectable increase in FISH spots above the background.
Combining both approaches yielded about 30% detection of transferred mRNA compared to the positive control.
This whole hide & seek story is summarized in the “Note 2” section and in Table 1 in the paper.
I learnt something new, but it does not feel like a win. That is because I detect less mRNA molecules than normal FISH, and only slightly better preservation of nanotubes. The use of proteinase K will probably affect any FISH-IF staining to detect the co-localization of protein and mRNA.
What is the mysterious protein coating that becomes impervious to FISH probes when the proteins are cross-linked by glutaraldehyde?
We do not know.
It seems that this protein coats the mRNA already during travel through the nanotubes, because I could not detect the mRNA in nanotubes either.
Coming back to the live imaging, it could be that the bulky 24x MCP-GFP that sits on the mRNA prevents proper coating of the mRNA with this mystery protein, thus inhibiting the transfer.
We are working now on several fronts to try to determine which proteins are involved in mRNA transfer through nanotubes.
To be continued…



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in