Scaffold hopping and fragment growing from amodiaquine yields a new Nurr1 agonist demonstrating potent target engagement and phenotypic effects in cellular PD model
Published in Chemistry, Cell & Molecular Biology, and Pharmacy & Pharmacology
Nuclear receptor related 1 (Nurr1, NR4A2) is a ligand-activated transcription factor expressed primarily in neurons and immune cells of the brain1–3. It functions as a monomer, homodimer or heterodimer and exhibits constitutive transcriptional activator activity even in the absence of ligands4,5. The dopamine metabolite 5,6-dihydroxyindole (DHI)6, polyunsaturated fatty acids7and prostaglandins A and E8 have been identified as endogenous Nurr1 ligands and recent efforts have yielded a few synthetic Nurr1 ligand chemotypes, of which the antimalarial drug amodiaquine (AQ) was the first validated direct Nurr1 agonist9. Nurr1 plays a critical role in regulating the development and maintenance of dopaminergic neurons and in brain inflammatory processes2,3,10,11. Observations of reduced Nurr1 levels in patients with Alzheimer's disease (AD) and Parkinson's disease (PD), as well as in corresponding animal models, suggest Nurr1 activation as promising approach in the treatment of neurodegenerative diseases12,13. Additionally, Nurr1’s relevance extends beyond the brain with a protective role in retinal pigment epithelial cells and therapeutic potential in age-related macular degeneration14. While AQ has been instrumental in early studies on the therapeutic impact of Nurr1 activation and revealed promising effects, the antimalarial is not specific for Nurr1 but exhibits multiple cellular effects including autophagy inhibition, p53 stabilization, suppression of ribosome biogenesis, and induction of endoplasmic reticulum stress15. Thus, confident target validation of Nurr1 with high-quality chemical tools is still pending11. To aid further pharmacological studies on Nurr1 with highly annotated chemical tools, we have developed potent and selective Nurr1 agonists from AQ focusing on rigorously validated activity and broad characterization.
Building on previous observations that the 7-chloroquinoline-4-amine substructure of AQ and chloroquine is sufficient for Nurr1 activation, we have probed alternative heterobicycles as Nurr1 ligands resulting in a scaffold hop to 8-chloro-2-methylimidazo[1,2-a]pyridin-3-amine as favored fragment-like Nurr1 agonist (Kd = 2.7 µM, EC50 = 7 µM). Subsequent fragment growing and systematic exploration of substituents in favored positions revealed opportunities for improving Nurr1 agonist potency. We obtained a high-affinity Nurr1 agonist (36, Kd = 0.17 µM, EC50 = 0.09 µM) exhibiting strong selectivity for NR4A over other nuclear receptors, favorable physicochemical properties and low toxicity. Additionally, we identified a structurally matched negative control compound (29) with no binding to Nurr1 to complement the agonist in biological studies. The agonist robustly induced Nurr1-regulated genes in T98G cells while the negative control had no effect demonstrating cellular target engagement and supporting suitability as chemical tool for in vitro experiments on the biology of Nurr1.
Next, we set out to apply the pair of agonist and negative control to a phenotypic experiment. We employed human midbrain organoids generated from induced pluripotent stem cells (iPSC) bearing a G2019S gain-of-function mutation in the leucine-rich repeat kinase 2 (LRRK2) gene or isogenic controls. LRRK2 mutation is among the most common genetic causes of Parkinson's disease and LRRK2 mutant organoids displayed diminished expression of Nurr1 and the dopaminergic neuron marker gene tyrosine hydroxylase (TH). Treatment with the agonist 36 rescued TH transcript levels of LRRK2 mutant organoids and enhanced the number of TH positive cells in both wild-type and mutant. The negative control 29 had no effect. These results provide further support for a promising role of Nurr1 as target in neurodegeneration and validate 36 and 29 as chemical tool.
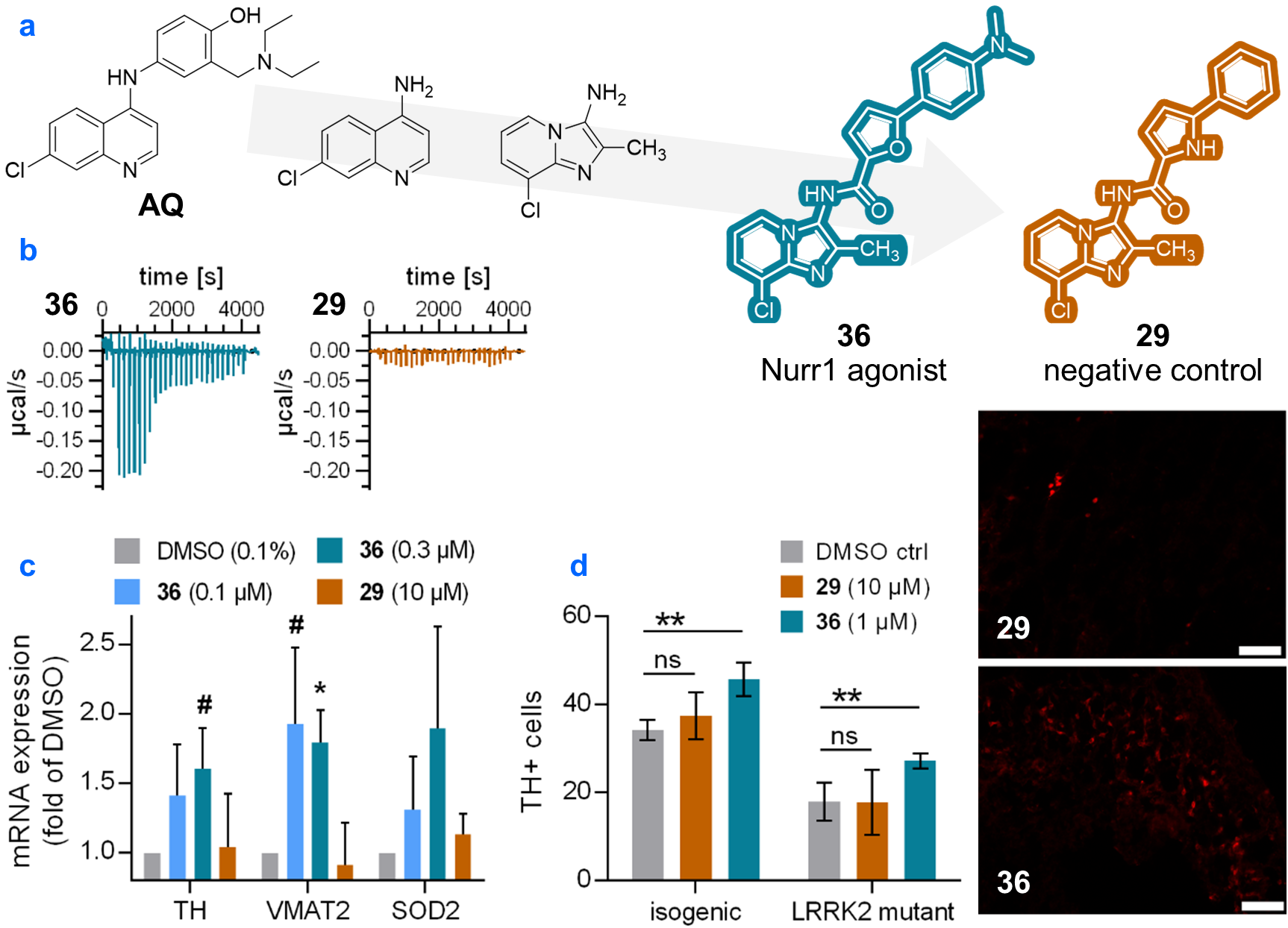
In summary, we report a new Nurr1 agonist that has been extensively validated for direct and cellular target engagement. Together with the structurally matched negative control, this agonist emerges as next-generation chemical tool to study the biology of Nurr1 in vitro.
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in