The diverse functions of bacterial purine receptors
Published in Microbiology
Receptor super-families that bind amino acids or biogenic amines
Typically, receptors are transmembrane proteins that contain an extracytosolic sensor domain and a cytosolic output domain. Sensor domains evolve very fast, as a consequence of which the nature of the ligand recognised is frequently not reflected in overall sensor domain sequence similarity. In general, the function of a bacterial protein can be inferred by overall sequence similarity with characterised homologues. However, due to this sequence variation this approach can only rarely be used to predict the signals recognised by sensor domains. We have solved the 3D structures of the PctA, PctB and PctC chemoreceptors from Pseudomonas aeruginosa that bind different amino acids. Although their sensor domains differ in sequence, the sensor domain residues that establish interactions with the bound ligand were highly conserved. Our close collaborator Igor Zhulin (Ohio State University) thus suggested that we should join forces to see whether the nature of receptor signals can be inferred from motifs of amino acids that interact with the bound signal.
Igor Zhulin is a computational biologist with extensive experience in the field of bacterial signal transduction. The dCache domain is the predominant bacterial extracytosolic sensor domain and present in all major receptor families. Based on 3D structures of dCache sensor domains containing bound amino acids, his team identified a conserved 5–amino acid motif that interacts with the bound amino acid signal. Sequence alignments showed that this motif was not conserved in dCache domains that bind other compounds like organic acids, polyamines, quaternary amines or purines. Database searches with this motif resulted in the retrieval of more than 10,000 sequences from bacteria, archaea and also human, that form the amino acid specific dCache_1AA family. However, do these domains indeed bind amino acids? To assess this issue we selected 8 domains, produced the recombinant proteins and conducted binding studies, showing that indeed all domains bound different amino acids (1). Receptors containing dCache_1AA domains were identified in all major bacterial receptor families. The amino acid responsive domains only correspond to a relative small fraction of dCache domains. Can thus further functional dCache domain families be defined? Using a similar strategy, we defined the 13,000-member dCache_AM family, that comprises receptors that bind different quaternary and biogenic amines (2).
Definition of the purine sensing receptor superfamily
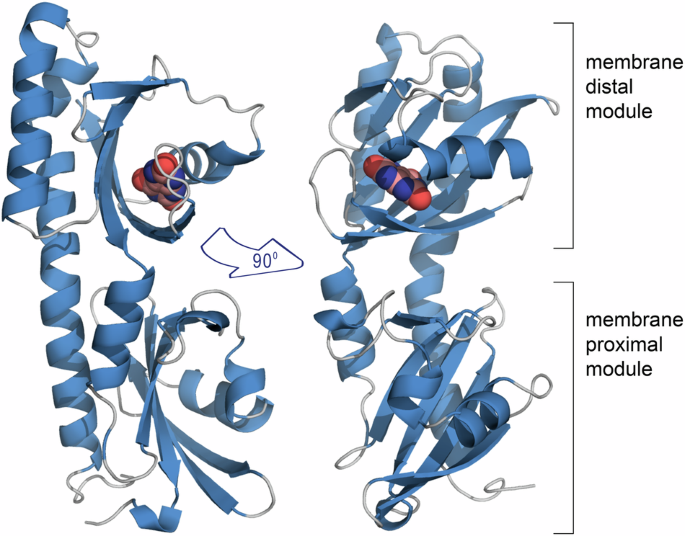
Amino acids and biogenic amines are known to be bacterial signal molecules. Purines are important signal molecules in human controlling different processes, but there is only scarce evidence for a role of purines as signal molecules in bacteria. When we identified the McpH chemoreceptor of Pseudomonas putida as the first purine specific receptor, the question was thus whether purine responsive receptors were just a little populated family present in bacteria with a specific lifestyle like the saprophytic P. putida, or whether there is a super-family of purine-responsive receptors, indicating a major role of purines as signal molecules. In the 3D structure of the McpH sensor domain (Figure), solved by our long-term collaborator Jose A. Gavira (Laboratory of Crystallographic Studies, Granada, Spain), bound uric acid (a purine) was mostly recognized by pi-stacking interactions with two aromatic residues, a binding mode very different to that of amino acids or biogenic amines.
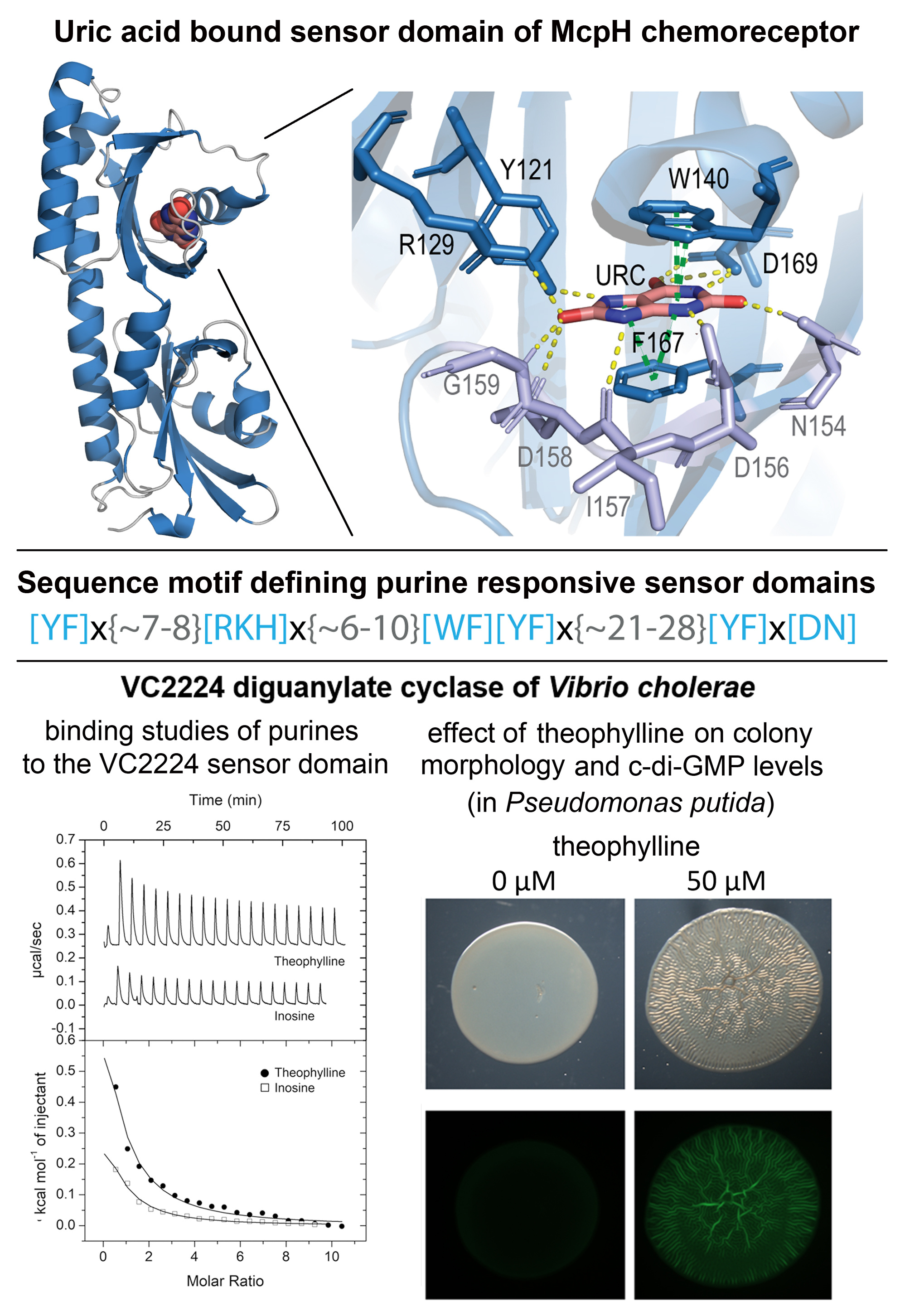
Based on the amino acids that are involved in uric acid binding to McpH, Igor Zhulin’s group defined a consensus motif (Figure). Searching sequence databases for dCache domains with this motif resulted in the retrieval of 6,300 proteins, forming the dCache_1PU purine responsive domain family.
Purine sensor domains are found in many different receptor families
The inspection of the members of the dCache_1PU family showed that purine sensor domains are not only found in chemoreceptors like McpH, but in other receptor families like sensor histidine kinases, serine/threonine phosphatases, diguanylate cyclases and phosphodiesterases that regulate a variety of processes like gene expression, metabolism, chemotaxis or second messenger levels. Among the strains harbouring purine responsive receptors were numerous human and plant pathogens. However, the question remained whether our predictions were correct. To assess this issue, we selected 15 proteins covering phylogenetic spread and different receptor families. Eleven proteins expressed and could be obtained as stable proteins permitting experimentation. Ligand screening showed that all 11 proteins bound purines and some of them pyrimidines (Note: purines are composed of a pyrimidine and an imidazole ring), confirming our prediction.
The enigma of theophylline
Adenine/adenosine and guanine/guanosine are products of nucleic acid degradation and thus abundant in many different ecological niches. We hence expected that purine sensors respond primarily to these compounds. However, we were wrong there since the analysis of the 11 proteins showed that all bound theophylline, whereas only few proteins bound purines derived from nucleic acid degradation. What is theophylline? It is a bimethylated purine, abundantly present in the tea plant and the active compound of tea. The analysis of metabolomics data showed that theophylline is found in all kingdoms of life. Interestingly, previous studies have identified an aptamer, widely exploited in synthetic biology applications, that binds specifically and with high affinity theophylline. Taken together, data thus suggest that theophylline is an important bacterial signal molecule. However, we are currently unable to identify the physiological significance.
Purines modulate c-di-GMP levels of a Vibrio cholerae diguanylate cyclase in vivo
One of the proteins that we have chosen for the experimental verification was the sensor domain of the VC2224 diguanylate cyclase from the human pathogen V. cholerae. Using microcalorimetry we showed that it bound the purines theophylline, guanosine and inosine (Figure). To assess the regulatory activity of the ligands in vivo, the receptor was expressed in P. putida KT2440, known for its low basal c-di-GMP level, harbouring a fluorescence-based c-di-GMP biosensor. As shown in the Figure, the addition of theophylline induced a wrinkly colony morphology, which was caused by the VC2224 mediated increase in c-di-GMP (green colour in the lower panel of the Figure). Ligands identified by in vitro experimentation thus possess the capacity to modulate receptor function in vivo.
Take-home message
Purines are important bacterial signal molecules that regulate a variety of cellular processes like gene expression, metabolism, second messenger levels or chemotaxis. Dedicated purine sensor modules have evolved to recognize specifically purines. Sensor domain ligands can be precisely predicted by motifs of amino acids that are involved in signal binding.
References
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in