The genetic landscape of autism spectrum disorder in an ancestrally diverse cohort
Published in Neuroscience and General & Internal Medicine
Who are we?
The Chahrour Lab at the University of Texas Southwestern Medical Center focuses on understanding the genetics and mechanisms underlying autism spectrum disorder (ASD) and related neurodevelopmental disorders. Our team of researchers, in collaboration with clinicians, employs techniques such as whole exome and genome sequencing to identify genetic variants associated with ASD. Through these discoveries, we aim to pave the way for innovative therapies and improve the lives of individuals affected by these disorders.
The Genetic Complexity of ASD
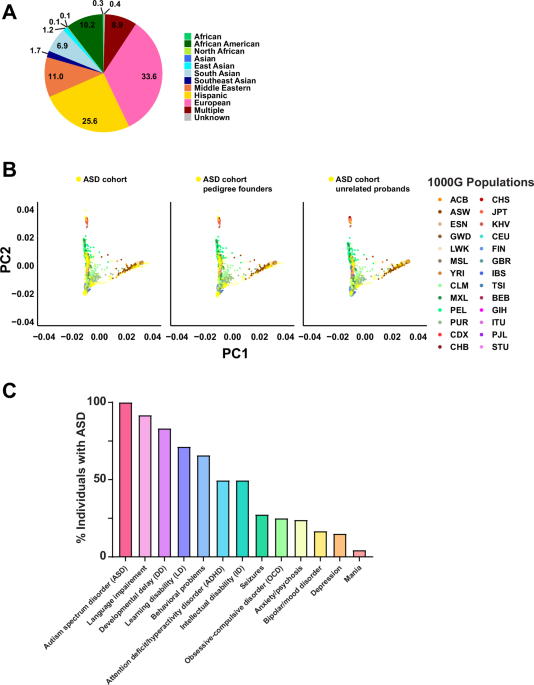
ASD is a collection of neurodevelopmental disorders characterized by challenges in social interaction, communication, and repetitive behaviors. Individuals with ASD may also exhibit a wide range of additional symptoms, including intellectual disability, anxiety, depression, attention disorders, and epilepsy. Recent estimates from the CDC indicate that approximately 1 in 36 children in the United States is diagnosed with ASD. While genetic factors play a significant role, accounting for about 80% of ASD cases, the specific genetic causes remain largely unknown. Each individual's unique presentation of ASD is influenced by a complex interplay of genetic and environmental factors.
Our Approach: A Diverse Cohort
We recruited a diverse cohort of 754 individuals from 195 families, each with at least one member diagnosed with ASD, representing over ten different ancestral backgrounds, including African American, Asian, Hispanic, Middle Eastern, Native American, and European. The ancestral diversity of our cohort distinguishes our study from many others, which often primarily focus on individuals of European ancestry. This initiative marks the first of its kind in the North Texas region to recruit families and conduct comprehensive genetic research focused on ASD.
Key Findings
Our comprehensive genetic analysis involved the development of variant filtering and prioritization pipelines to pinpoint potential causal variants in individuals with ASD. This analysis has yielded several important findings:
Identification of Novel Variants: We discovered 38,834 novel genetic variants that have not been previously reported in any population database.
Diagnostic Yield: Notably, we achieved a diagnostic rate of 30%, identifying 92 potentially pathogenic variants in 73 known ASD genes, such as BCORL1, CDKL5, CHAMP1, KAT6A, MECP2, and SETD1B.
Discovery of Novel Candidate Genes: We have identified 158 potentially pathogenic variants in 120 candidate genes that may play a role in ASD and require further functional studies, including DLG3, GABRQ, KALRN, KCTD16, and SLC8A3.
Convergence of Implicated Pathways: We found that the identified genes function primarily in nervous system development and regulation of signal transduction.
Conclusion
By sequencing a diverse cohort of individuals with ASD from over ten ancestries, this study breaks away from the limitations of single-population analyses and contributes to the ongoing effort to identify causative genes and variants. To learn more about our research or to join our study, please visit https://chahrourlab.org/.
Follow the Topic
-
npj Genomic Medicine

This is an international, peer-reviewed journal dedicated to publishing the most important scientific advances in all aspects of genomics and its application in the practice of medicine.
Related Collections
With Collections, you can get published faster and increase your visibility.
Artificial Intelligence in Genomic Medicine
Publishing Model: Open Access
Deadline: Jun 23, 2026
Genomics in precision medicine
Publishing Model: Open Access
Deadline: Apr 14, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in