The Identification And Characterization Of A Selected SAM Dependent Methyltransferase Ribozyme That Is Present In Natural Sequences
Published in Chemistry
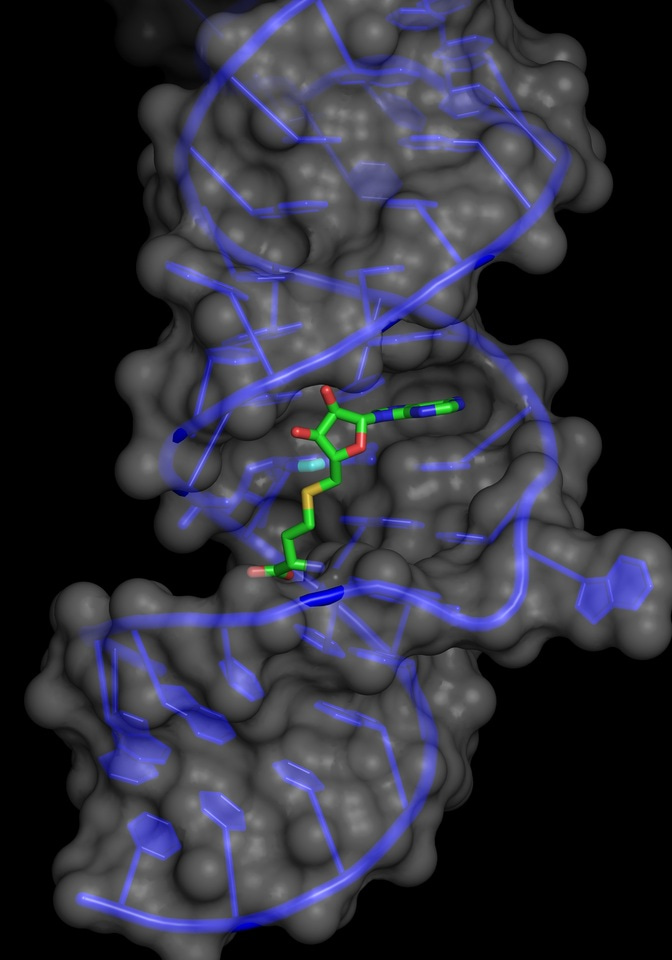
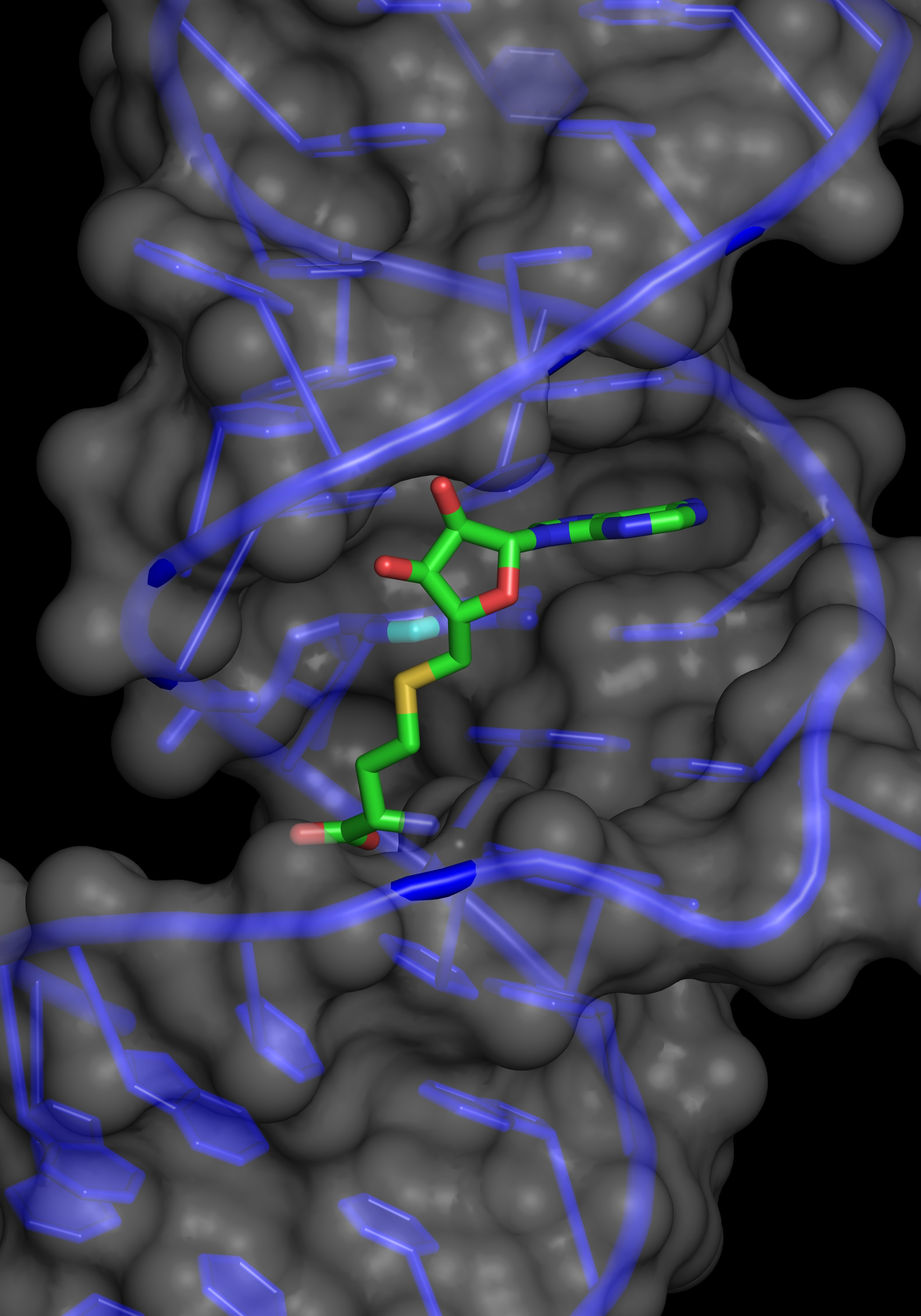
For some time, it has been known that cellular nucleic acids including both DNA and RNA molecules are chemically modified. The modifications involve chemical changes at positions in the nucleosides. Such modifications play a central role in the emerging disciplines of epigenetics and epitranscriptomics. Over 100 modifications to cellular RNAs including mRNAs and SnoRNAs have been identified 1. The RNAs of the translational machinery: rRNA and tRNA are extensively modified. The ubiquity of the translational machinery means that cells devote considerable resources to the modification of tRNAs and rRNAs. Although a number of modifications are possible, the majority involve the addition of a methyl group to positions within the nucleoside.
Specific methyltransferase enzymes transfer the methyl group from the cofactor, S-Adenosyl-L-methionine (SAM), to the nucleic acid. The methyl transferase enzymes bind SAM 2 and orient the highly reactive sulphonium cation of SAM for methyl group transfer.
Ribonucleotide cofactors are essential in all cells, and were probably retained from the ‘RNA world’ by evolution. The ribonucleotide cofactors from the RNA world would later be recruited as cofactors by protein enzymes 3. Of the enzyme cofactors only ATP exploits a greater variety of enzymes than SAM 4,5. The N7 position of guanine is the most reactive position on DNA for chemical methylation by SAM 6.
Natural ribozymes that are involved in essential cellular processes such as translation and splicing, appear to have been retained from the RNA world by evolution 3. Over time ribozymes that catalyse other types of metabolic reactions appear to have been surpassed by protein enzymes. A number of ribozymes have been selected by Systematic Evolution of Ligands by EXponential enrichment (SELEX) in vitro, to perform simple chemical and biological reactions. Recently, m6G and O6-methyl pre-queuosine dependent methyltransferase ribozyme activities have been reported 7,8. However, no methyltransferase ribozyme RNA that used SAM as a cofactor has been identified.
Previous studies had shown that methyltransferase enzyme activity could be sensitive to the conformation of DNA and RNA substrates 9,10. Methyltransferase proteins encapsulate SAM in an inert environment such that the reactive methyl-group becomes available for methylation upon RNA binding 11. Similarly, the riboswitch RNAs that bind SAM to regulate aspects of the translation and transcription of SAM metabolism also encapsulate SAM in an unreactive environment 12.
From this we reasoned that the riboswitch RNAs that bind SAM might be possible candidate molecules to investigate (protein enzyme-free) methyl transfer from SAM to RNA. For riboswitches I-IV, we explored a range of reaction conditions including incubation time, temperature, pH, metal ions and organic solvents for methylation. Using [H3 SAM] as a methyl donor, low levels of [H3] were detected in riboswitch RNAs in the presence of copper ions. A combination of mass spectrometry and HPLC confirmed the presence of m7G in the RNA. This was encouraging, although the levels of [H3] incorporation were relatively small compared to protein methyltransferases. These experiments had enabled us to ‘road-test’ the methodology for assaying protein-free RNA methylation and gave us a baseline from which we could try to identify RNA molecules that could self-methylate.
To identify RNA molecules that could efficiently self-methylate, we used SELEX. A specific RNA sequence capable of self-methylation was selected using an m7G specific antibody to enrich the modified RNA sequences for each round of SELEX.
Typically, successful SELEX experiments require more than 12 cycles to identify good ligands, but after only 6 rounds of SELEX, high levels of [H3] from [H3 SAM] were incorporated into the RNA which was a good sign. To rule out the possibility that the modification was random and not specific, a selection of sequences were tested for methylation activity and an active sequence identified. This could be shortened to a 33-nucleotide fragment with methyl transfer activity which we dubbed SMRZ-1.
The active SMRZ-1 RNA sequence was G rich and HPLC and mass spectrometry confirmed the presence of m7G in the RNA. Further biochemical analysis confirmed that the RNA behaves as a methyltransferase enzyme that modifies a specific G nucleotide in the RNA and can be characterised as an enzyme. Characterisation of the ribozyme showed it to offer a number of distinct points of difference compared to a protein methyltransferase.
The short sequence of the ribozyme made it an attractive candidate for structural studies. The structure of the RNA was determined both with and without SAM, surprisingly, in the absence of SAM the RNA adopts a novel motif, that undergoes a conformational change upon SAM binding 13.
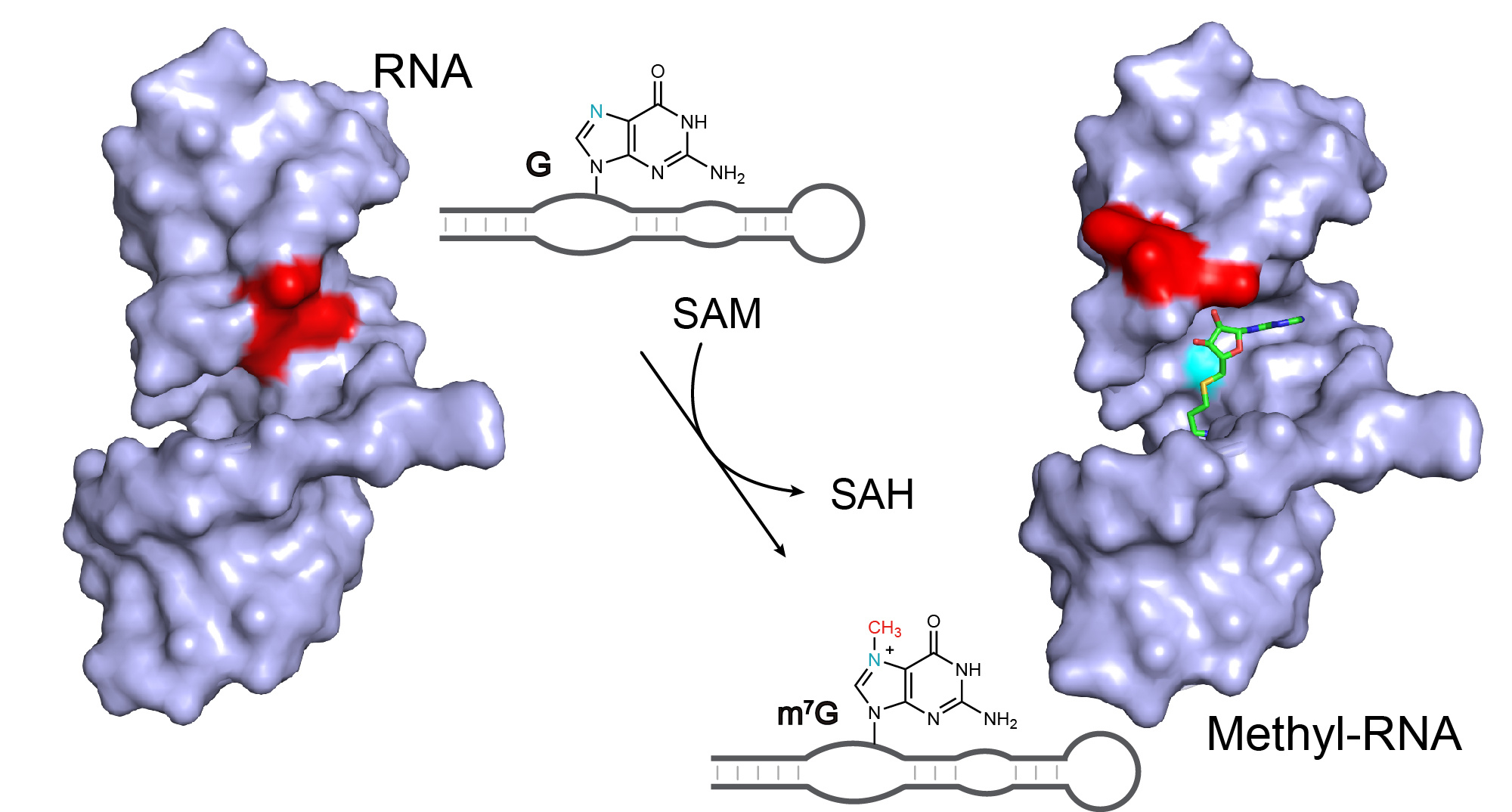
Mutagenesis and structural analysis identified essential nucleotides in the core of the ribozyme. Bioinformatic searches identified a number of natural sequences that were active in vitro with a range of activities. Such sequences contain the original core nucleotides but differ elsewhere in the molecules. Potential ribozyme sequences were identified in a range of organisms. We anticipate that the continuing expansion of genomic data and improved searching will identify additional ribozyme candidates. The SMRZ-1 ribozyme has optimum activity at >65°C in the presence of Cu (II) and ribozyme candidate sequences are enriched in thermophilic strains. The biochemical properties of the SMRZ-1 ribozyme may be particularly suited to organisms that grow in environments that experience fluctuations in pH, ionic strength and temperature.
The importance of specific RNA modifications in the modulation of gene expression is becoming increasingly apparent. A co-transcriptional methyltransferase ribozyme would offer some benefits to the cell; e.g., the circumvention of the protein synthesis machinery to methylate RNA might provide a fitness benefit under certain growth or developmental conditions. The identification of the SMRZ-1 ribozyme offers some support to the notion that other similar methyltransferase ribozymes may have been active in the RNA world. The existence of ribozyme sequences in natural sequences also opens up the possibility that other methyltransferase ribozymes may contribute to cellular methylation. Such ribozymes have yet to be identified but they are an intriguing and exciting prospect.
Related Content:
https://www.nature.com/articles/s41929-021-00685-z
https://rdcu.be/czSLo
Alastair I. H. Murchie, Dongrong Chen, Jianhua Gan, Hengyi Jiang, Yanqing Gao and Lei Zhang
References
- Benne, R. & Grosjean, H. Modification And Editing Of RNA. (American Society for Microbiology, 1998).
- Schubert, H. L., Blumenthal, R. M. & Cheng, X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci 28, 329–335 (2003).
- Benner, S. A., Ellington, A. D. & Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. U.S.A. 86, 7054–7058 (1989).
- Cantoni, G. L. Biological methylation: selected aspects. Annu. Rev. Biochem. 44, 435–451 (1975).
- Loenen, W. a. M. S-adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 34, 330–333 (2006).
- Rydberg, B. & Lindahl, T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J 1, 211–6 (1982).
- Scheitl, C. P. M., Ghaem Maghami, M., Lenz, A.-K. & Höbartner, C. Site-specific RNA methylation by a methyltransferase ribozyme. Nature 587, 663–667 (2020).
- Flemmich, L., Heel, S., Moreno, S., Breuker, K. & Micura, R. A natural riboswitch scaffold with self-methylation activity. Nat Commun 12, 3877 (2021).
- Murchie, A. I. & Lilley, D. M. Base methylation and local DNA helix stability. Effect on the kinetics of cruciform extrusion. J Mol Biol 205, 593–602 (1989).
- Yin, S., Jiang, H., Chen, D. & Murchie, A. I. H. Substrate recognition and modification by the nosiheptide resistance methyltransferase. PLoS ONE 10, e0122972 (2015).
- Yang, H. et al. Crystal structure of the nosiheptide-resistance methyltransferase of Streptomyces actuosus. Biochemistry 49, 6440–6450 (2010).
- Price, I. R., Grigg, J. C. & Ke, A. Common themes and differences in SAM recognition among SAM riboswitches. Biochim. Biophys. Acta 1839, 931–938 (2014).
- Jiang, H. et al. The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences. Nat Catal 4, 872–881 (2021).
Follow the Topic
-
Nature Catalysis

This journal brings together researchers from across all chemistry and related fields, publishing work on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, incorporating both fundamental and applied studies.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in