Unveiling the Mystery of Soil Microbial Dark Matter
Published in Microbiology

In a groundbreaking revelation, a single gram of surface soil has been found to harbor billions of bacterial and archaeal microorganisms along with a staggering trillions of viruses. This astonishing revelation underscores the previously underestimated richness of soil microbial diversity, which surpasses that of most other environments for its remarkable complexity and spatial heterogeneity.
Despite this wealth of microbial life, previous studies have largely skirted the challenges posed by the intricate metagenomes of soil, which are brimming with genomes from uncultivated and enigmatic microorganisms. The majority of soil microbiome research has been hampered by limitations and biases inherent in reference databases, making it difficult to characterize microbes with the desired taxonomic precision.
While some studies have managed to recover genomes from soil metagenomes on a smaller scale and across multiple systems, the vast reservoir of soil metagenomes available in public databases remains largely untapped on a global scale.
In a pioneering effort, we have embarked on a mission to establish a comprehensive public resource database and delve into the enigmatic realm of soil microbial dark matter. Our first step involved the reconstruction of Metagenome-Assembled Genomes (MAGs) from metagenomic datasets spanning the globe. This ambitious endeavor aims to significantly expand the genomic catalogue of soil microbiomes, shedding much-needed light on the mysteries of microbial dark matter lurking beneath the earth's surface.

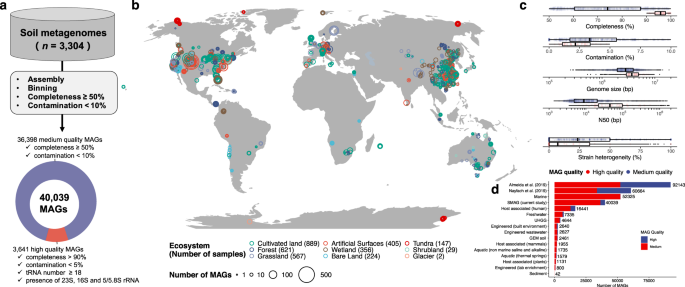
We conduct the first large-scale excavation of soil microbial dark matter by reconstructing 40,039 metagenome-assembled genome bins (the SMAG catalogue) from 3,304 soil metagenomes (Fig.1). By aligning SGBs with approximately 500,000 reference genomes from the Refseq database and MAGs from other studies, we have identified an astounding 16,530 unknown SGBs (uSGBs). In-depth analyses, including intraspecific pangenome profiles and single nucleotide variants (SNVs), have unveiled the vital functional roles played by these uSGBs within soil microbiomes. Our exploration didn't stop there. We ventured into the realm of Biosynthetic Gene Clusters (BGCs) and CRISPR-Cas genetic resources, confirming the immense potential harbored within soil microbiomes for mining genetic resources. Furthermore, we have unearthed previously concealed viral-host associations residing within the MAGs, opening up new avenues of research in this dynamic field.
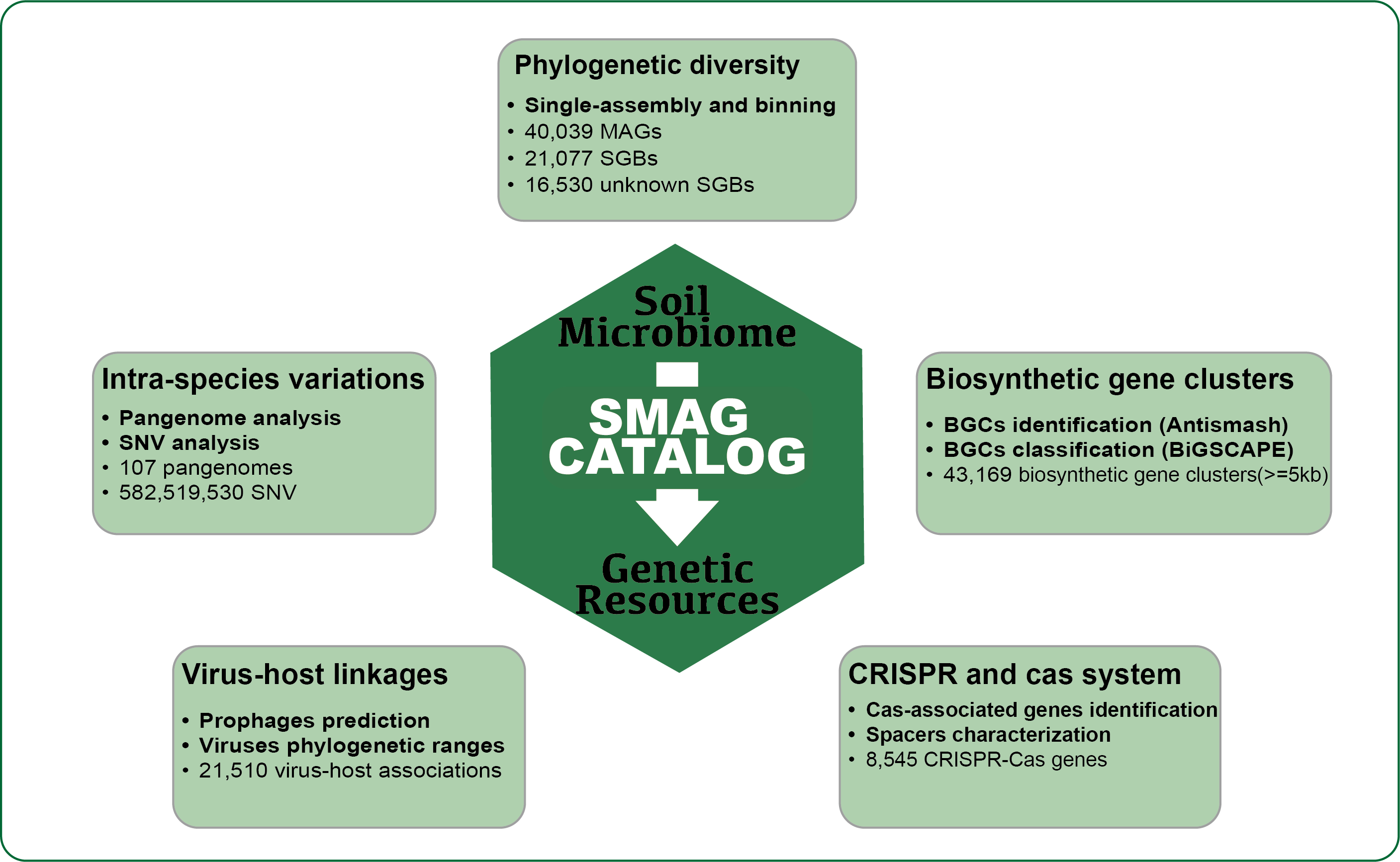
The Soil Metagenomic-Assembled Genome (SMAG) catalogue constitutes abundant information, offering invaluable opportunities for future studies aimed at unraveling the ecological roles of soil microbiomes and identifying genetic resources that could serve as an important resource for future broad innovations.
Detailed information can be acquired through https://smag.microbmalab.cn.
Acknowledgements
Sincerely, we thank C. Kelly, C. Averill, D. Buckley, D. Goodheart, D. Duncan, D. Myrold, E. Eloe-Fadrosh, E. Brodie, E. H.gfors-R.nnholm, H. Cadillo-Quiroz, J. Tiedje, J. Jansson, J. Norton, J. Blanchard, J. Schweitzer, J. Banfield, J. Gladden, J. Raff, K. Peay, K. Gravuer, K. M. DeAngelis, L. Meredith, M. Kalyuzhnaya, M. Waldrop, N. Fierer, P. Dijkstra, P. Baldrian, S. Theroux, S. Tringe, T. Woyke, T. Whitman, W. Mohn & San Diego State University for their permission to use their metagenome data.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in