A database on the abundance of environmental antibiotic resistance genes
Published in Earth & Environment
Explore the Research
A database on the abundance of environmental antibiotic resistance genes - Scientific Data
Scientific Data - A database on the abundance of environmental antibiotic resistance genes
Background
Antimicrobial resistance (AMR) has raised global public health concerns. A global study1 has estimated that AMR contributed to about 1.27 million direct deaths and 4.95 million indirect deaths in 2019. A review2 has predicted that AMR could cause 10 million deaths each year by 2050. It’s significant for us to take prevention and control measures to guard against such a threat. What can we do in detail? Tracking the occurrence and prevalence of antibiotic resistance genes (ARGs) is a key step.
The wide distribution of ARGs in the environment contributed significantly to AMR. ARGs can persist for a long time in the environment and can be transferred within the microbial community. Note that more and more bacterial species have had the capacity to tolerate antibiotics over the years. With the aid of mobile genetic elements (MGEs), ARGs can be horizontally transferred from environmental antibiotic-resistant bacteria to clinical pathogens, further exacerbating the risks to human health. Consequently, the risk assessment of ARGs has become a global urgence, where the primary step is to monitor and profile the occurrence and distribution of ARGs across the environment.
As emerging contaminants, ARGs have been globally detected in various habitats. However, few databases on the surveillance data of the spatiotemporal distribution and abundance of environmental ARGs have been developed, especially on the absolute level. A high-throughput quantitative PCR (HT-qPCR) method, which can achieve 5184 times of PCR reaction in a plate one time, was established in our lab and widely used by global researchers. This method has the advantages of better detection limits, lower cost, reducing sample quantity requirement, and absolute quantification. With this technology, large amounts of data about the levels of ARGs in the environment were produced in the past ten years. Part of the data was further analyzed and published in Nature Microbiology, and other high impact journals.
Data generation
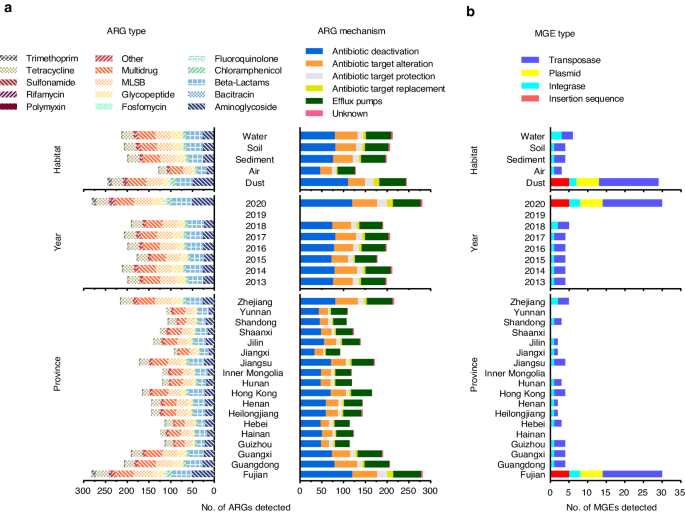
In this study, the data were obtained from our and the collaborator’s laboratories. We compiled the ARG occurrence data from a total of 1403 samples in 653 sampling sites across 18 provinces in China from 2013 to 2020. All the data were uniformly generated using high-throughput quantitative PCR under the same standard operational protocols, thus showing high comparability within samples. We reclassified the ARGs into 15 types according to the antibiotics to which they conferred resistance. Also, we divided them into six groups based on the mechanism of resistance: antibiotic deactivation, efflux pumps, antibiotic target alteration, antibiotic target protection, antibiotic target replacement, and unknown. In addition, the health risk of ARGs was categorized into four ranks according to a previous study3.
Introduction of the database
The database possessed 291,870 occurrence records from five types of habitats (water, soil, sediment, dust, and atmosphere) on the abundance of 290 ARGs and 8,057 records on the abundance of 30 mobile genetic elements (MGEs). It consists of two separate tables, providing the abundance of ARGs and MGEs with detailed gene information and sample information, respectively. For gene information, the resistance type, resistance mechanism, and health risk rank were introduced to classify different ARG subtypes. The MGE type is also provided. For sample information, sampling date, sampling location, and habitat are provided. In addition, the information about the publications using the data in the database is provided.
Benefits of the database
To the best of our knowledge, this is the first database that provides large amounts of ARG spread data including both absolute and relative abundance data. The database provides a valuable resource for exploring the patterns of antibiotic resistance and studying the transfer of ARGs facilitated by MGEs. The spatial distribution analysis of ARGs can be used to identify microbial resistance hotspots. The database also provides basic information for studying the health risk assessment of AMR.
Data availability
The data can be downloaded in the form of Excel spreadsheets (.xlsx) from https://doi.org/10.57760/sciencedb.09803. We also developed an online platform (http://sdc.iue.ac.cn/abundance/arg/list)to show the data. It has a friendly interface and provides the search to obtain the profiles and abundance of ARGs in a specific period and place.
We hope to incorporate new ARGs and more abundance data from various habitats in the future, allowing for a broader spatial and temporal analysis.
References:
- Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629-655 (2022).
- O'Neill, J. Tackling drug-resistant infections globally: Final report and recommendations. (Review on Antimicrobial Resistance, 2016).
- Zhang, A.-N. et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 12, 4765 (2021).
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Invertebrate omics
Publishing Model: Open Access
Deadline: May 08, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in