Artificial cells compete for proteins
Published in Chemistry, Materials, and Cell & Molecular Biology
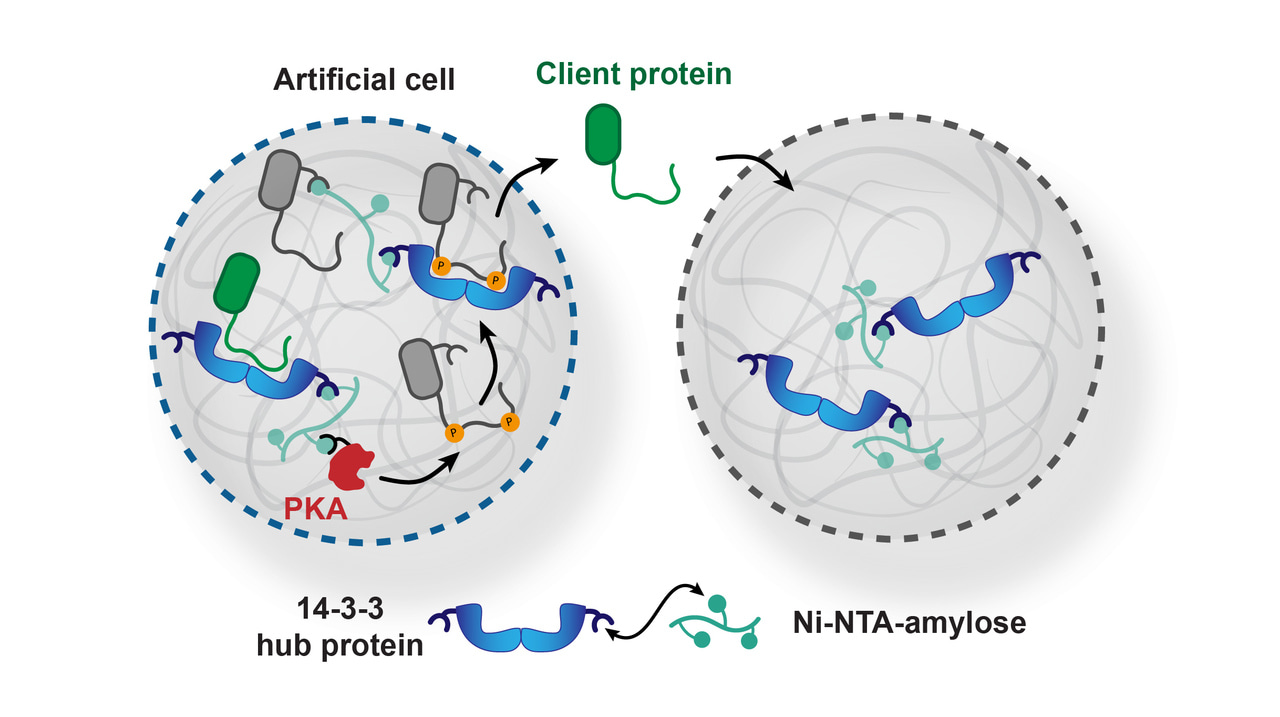
Living cells can communicate with each other by the exchange of soluble molecules such as hormones or growth factors. These soluble molecules can be released by cells and diffuse to a different population of cells, where they can bind to receptors on the surface and/or be internalized. As there are many different cell types and environments within our bodies, the effect of such signaling processes needs to be controlled. Cells have therefore adapted their sensitivity and affinity for such signaling molecules, and they often compete with each other for their binding. To gain more insight in these competitive binding events a suitable model system would be very useful.
We wondered if an artificial cell system could be used to study such binding events. Bottom-up artificial cells, typically formed by biopolymers or lipids, offer a degree of control over the formulation and environment that is typically not achievable in cellular systems. Especially coacervates have often been used. They can be formed by the mixing of two oppositely charged polymers, which spontaneously yields a polymer-rich phase. This phase has a density which is similar to the molecular density found in living cells. In our lab, we have been working with an amylose-derived coacervate platform.1,2 We can specifically take up proteins of interest into these coacervates in high concentrations by interactions with the coacervate building blocks, reaching >100-fold enrichment over the bulk concentration. Usually, coacervates fuse on a short timescale within several hours to form a macrophase. Importantly, our coacervate formulation is stable over several weeks in solution due to the semipermeable membrane that is formed by a triblock copolymer that we designed for this purpose.
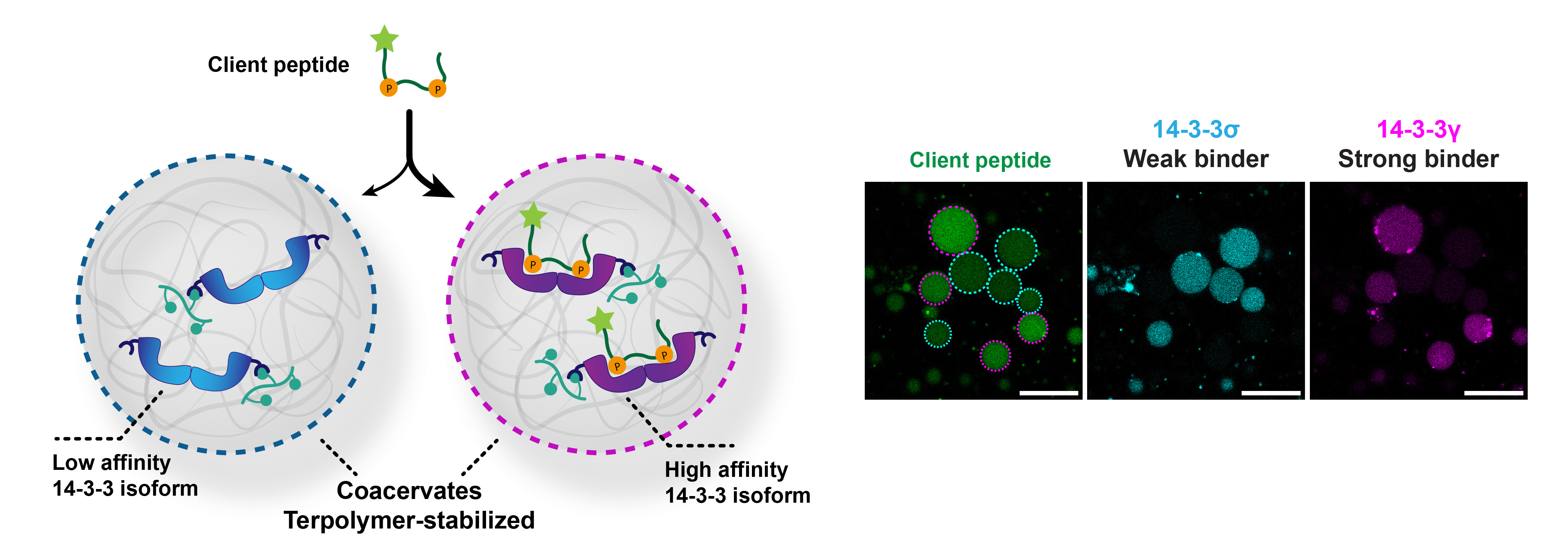
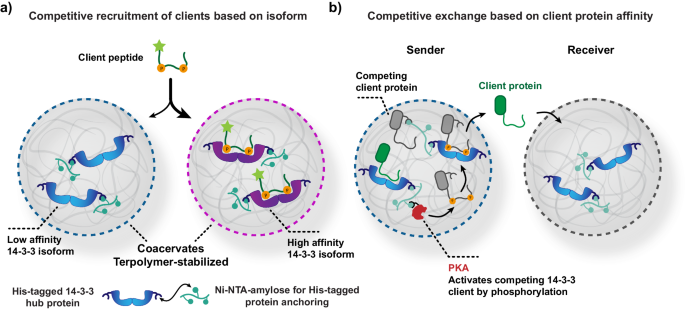
In our lab, we have a great deal of experience with the protein-protein interactions of the hub protein 14-3-3. Its rather cryptic name might sound like it is a niche protein – the name is derived from the experiment in which it was discovered in 1967 – but it is a protein involved in many key signaling pathways.3 Important 14-3-3 clients include the tumor suppressor protein p53.4 The 14-3-3 protein has seven different variants (isoforms) which have conserved binding grooves but differences in binding affinity to client proteins. We hypothesized that we could use the difference in affinity between two variants to direct a client of 14-3-3 to a specific population of coacervates.
We loaded one coacervate population with a low affinity 14-3-3 isoform, and a second population with a high affinity isoform (Figure 1). By confocal microscopy, we monitored the recruitment of a strongly binding client peptide. There were no significant differences in client recruitment between the individual samples of coacervates containing a single population. The local concentrations of 14-3-3 exceeded the KD values by several orders of magnitude. However, in a mixed coacervate population with excess 14-3-3 binding sites, we observed that the client was indeed selectively recruited to the coacervate population with the high affinity isoform. This demonstrates that competitive interactions can drive selective recruitment events.
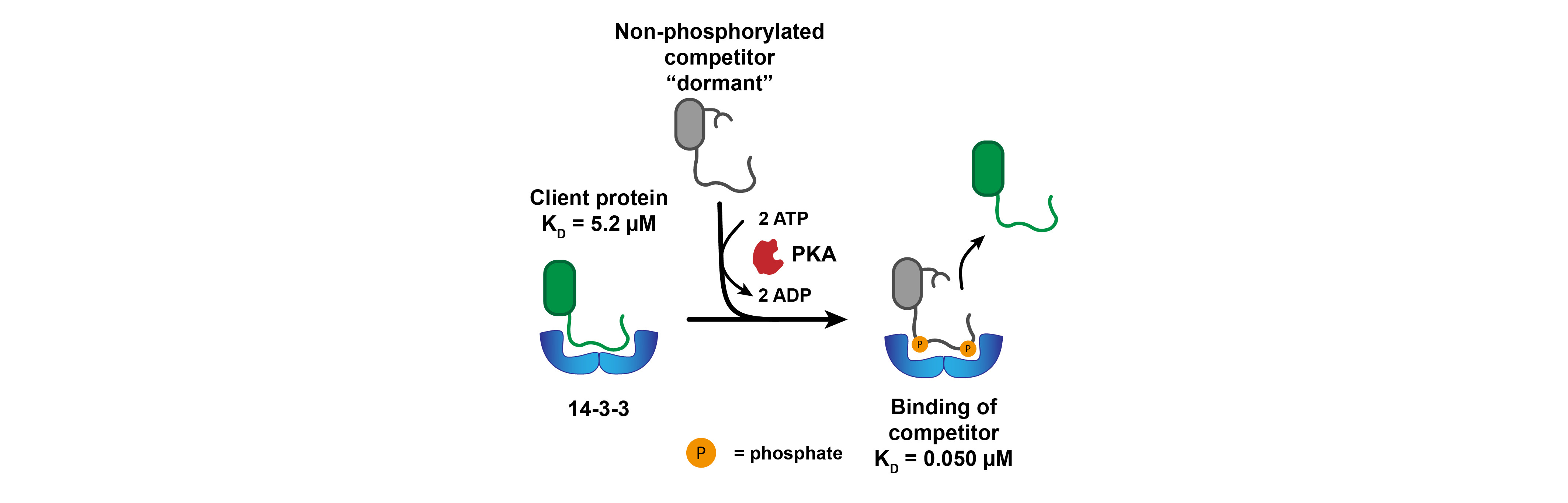
After demonstrating the competitive recruitment of a 14-3-3 client to coacervates based on 14-3-3 isoform, we wondered if we could gain control over such competitive interactions. We engineered a competitive binder for 14-3-3, that requires double phosphorylation to bind with high affinity. Upon phosphorylation by a kinase, the competitive binder displaced an initially bound client protein with moderate affinity (Figure 2).
Next, we used these competitive intracellular interactions to displace the client protein from coacervates with the strong isoform of 14-3-3. The client protein was then able to diffuse to coacervates that are loaded with the weak isoform – against the natural recruitment potential of these populations (Figure 3). The client protein was initially bound in the coacervates with the strongly recruiting isoform (sender). Upon phosphorylation of the intracellular competitor, the client protein was outcompeted. Subsequently, the client protein was recruited into a receiver population of coacervates containing a weakly binding isoform of 14-3-3. The release of the client protein was found to be surprisingly rapid, with near-complete release with 30 min. We believe this is due to the unfavorable environment that the highly charged coacervate provides for the nearly charge-neutral, well-hydrated client protein.
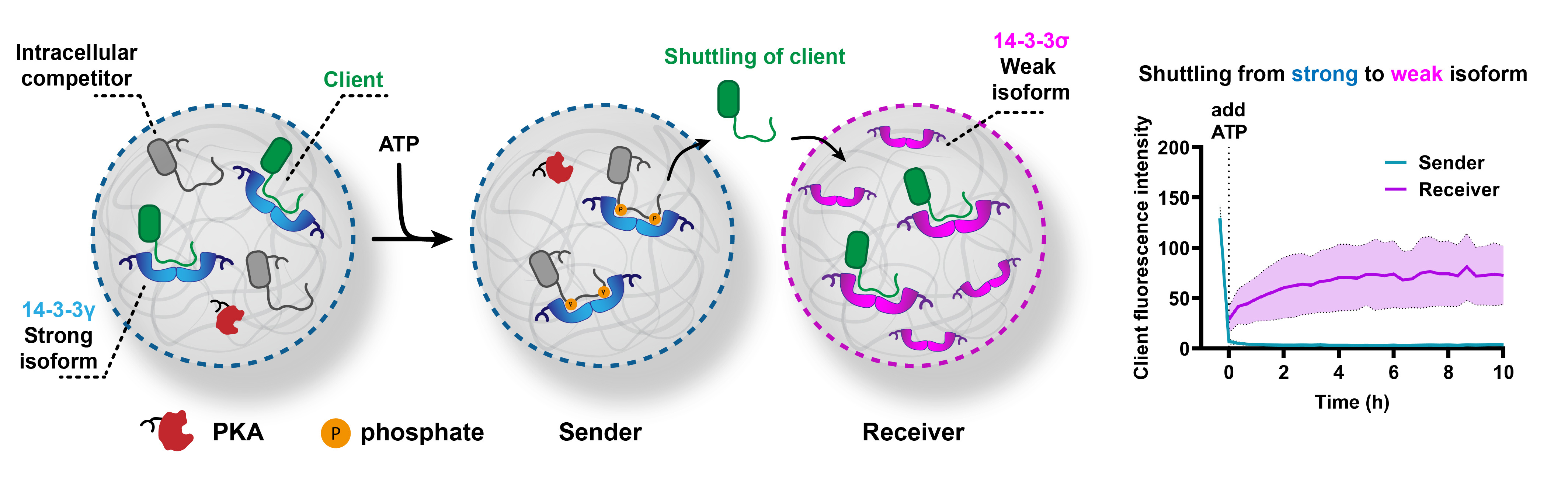
Proteins need to be retained over time in specific coacervate populations to study such systems, and this was challenging for us, since the coacervate environments are in equilibrium with each other and the bulk solution. We engineered stable recruitment of the proteins inside our artificial cells by enhancing the interactions with the coacervate building blocks, and this allowed us to retain the protein cargo in specific populations over time. Our results demonstrate that affinity engineering and competitive binding can provide directed protein recruitment and exchange between artificial cells.
In future work, we aim to expand this system by studying the exchange of functional proteins rather than model fluorescent proteins. One can think of many interesting targets involved in cellular signaling pathways, with a more complex readout driven by enzymatic activity for example. The competitive binding events between artificial cells may be used as a model system for cells with different affinity for soluble signals, such as distinct cell types or pathogenic cells, with overexpressed or mutated proteins.
References
- Altenburg, W. J. et al. Programmed spatial organization of biomacromolecules into discrete, coacervate-based protocells. Nat. Commun. 11, 6282 (2020).
- van Veldhuisen, T. W. et al. Enzymatic Regulation of Protein–Protein Interactions in Artificial Cells. Adv. Mater. 35, 2300947 (2023).
- Aitken, A. 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 16, 162–172 (2006).
- Rajagopalan, S., Sade, R. S., Townsley, F. M. & Fersht, A. R. Mechanistic differences in the transcriptional activation of p53 by 14-3-3 isoforms. Nucleic Acids Res. 38, 893–906 (2009).
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in