Unlocking mushroom treasures: the untold story for reconstructing the biosynthetic network of type II ganoderic acids in engineered yeast
Published in Cell & Molecular Biology, Pharmacy & Pharmacology, and Agricultural & Food Science
Mushrooms represent an abundant source of medically relevant natural products. They can be used as medicines for the ill, tonics (dietary supplements) for those in a sub-health state as well as for both healthy and ill individuals, and as foods for health people worldwide. In contrast to the vast global market of the mushroom industry and the importance of mushroom-derived compounds, related basic research has been neglected.
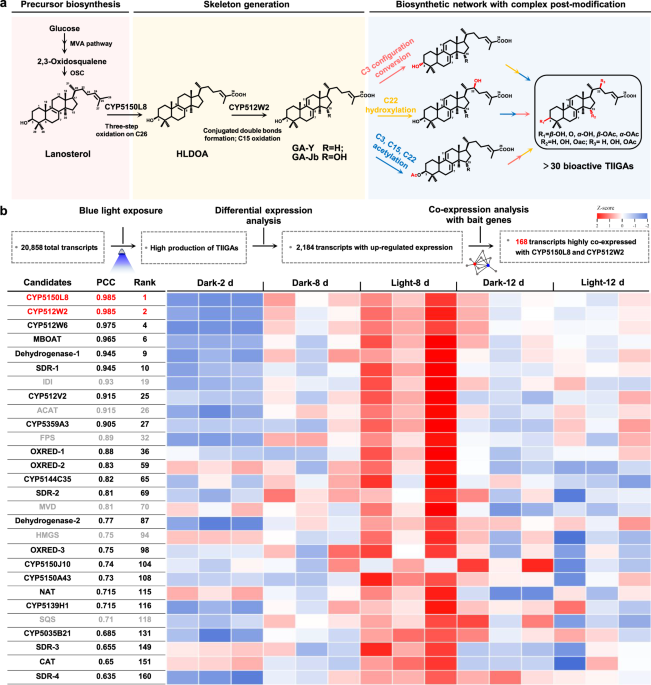
One remarkable example of mushrooms is the traditional Chinese medicinal Ganoderma lucidum. Its significant pharmacological properties are due to the dosage and composition of its specific active metabolites, lanostane-type triterpenoid type II ganoderic acids (TIIGAs). However, the inaccessibility of such active metabolites is the foremost challenge to the widespread application of Ganoderma products. Our laboratory has long been focusing on the analysis and reconstruction of the TIIGAs’ biosynthetic pathway, hoping to realize its efficient heterologous production through a synthetic biology platform to overcome the challenges of the immature genetic manipulation, the long growth cycle, and the susceptibility to environmental factors of G. lucidum.
After persistent efforts by senior researchers in our lab, two key cytochrome P450s (CYPs) were successfully identified as responsible for forming the basic skeleton of TIIGAs1,2. Prior to this study, we had systematically screened 158 CYPs from G. lucidum, representing all the coding CYPs we can obtain at that time. Despite this comprehensive screening, the critical C22 hydroxylase remained elusive2. Faced with this complete lack of leads, we formulated a working hypothesis: any newly obtained CYP from G. lucidum that catalyzes the production of a new fermentation product with a mass increase of 16 m/z compared to its most likely substrate, GA-Jb, could potentially be the missing C22 hydroxylase. After extensive optimization of PCR conditions, we successfully obtained one newly CYP and introduced it into the GA-Jb producing yeast. The transformant produced a novel product with an increased m/z of 16 compared to GA-Jb. This exciting discovery promoted us to devote nearly six months to thorough in vitro enzymatic characterization and NMR-based structural elucidation. However, contrary to our initial hypothesis, this enzyme was not the C22 hydroxylase, a finding that would ultimately lead our research in unexpected new directions. In our ongoing pursuit of the C22 hydroxylase, we implemented a multi-pronged strategies to (1) significantly increase the accumulation of TIIGAs during G. lucidum mycelial growth, (2) facilitate capturing more missing enzymes, and (3) enable chemical synthesis of C22 hydroxylated GAs as standards to direct our enzyme discovery efforts.
Co-expression analysis proved particularly valuable for identifying the most likely candidate enzymes, including CYP and acetyltransferase. Our successful identification of CYP512W6 as the C22 hydroxylase using the GA-Jb-producing yeast screening system inadvertently caused the mischaracterization of GlAT as a monofunctional C15 acetyltransferase, since GA-Jb lacked C22 hydroxylation. When we tested the substrate preference of GlAT on TLTOA, a C22 hydroxylated GA, we observed multiple reaction products. However, a comprehensive analysis of the LC-MS results was initially overlooked due to our priori assumption that a single acetyltransferase would be unlikely to catalyze acetylation at both C15 and C22 positions of the triterpenoid skeleton, given the considerable distance between these hydroxyl groups. Thus, we spent a long time attempting heterologous candidates of C22 acetyltransferase, yet all efforts proved unsuccessful. Our attention was subsequently drawn to the consistent production of multiple reaction products when GlAT was incubated with GA-T2. By carefully re-examined the LC-MS data, we ultimately confirmed the bifunctional acetyltransferase activity of GlAT at C15 and C22.
Inspired by these discoveries, we explored the catalytic mechanism of the C22 hydroxylase CYP512W6 and the C15 and C22 bifunctional acetyltransferases GlAT. Combining computational-aided design with experimental verification, we identified key residues critical for their catalytic activity and substrate selectivity, shedding lights on subsequent protein engineering. Following the introduction of all identified enzymes into the yeast system, we detected a significant metabolic bottleneck at the GA-TN intermediate during TIIGA biosynthesis. Based on our understanding of the catalytic functions of these enzymes, we adopted a temporal expression control strategy for the acetyltransferases BsAT and GlAT, which successfully alleviated the metabolic flux obstruction and restored production of the target product.
In summary, our work not only completely decoded the biosynthetic pathways of TIIGAs, but also reprogrammed it to target promising compounds through systematic engineering work. With the platform established in this work, we achieved de novo biosynthesis of over 30 bioactive TIIGAs, exhibiting 1 to 4 orders of magnitude higher titers or efficiencies than those from farmed mushrooms. It enables the development of functional foods and the in-depth pharmacological study of mushroom-specified bioactive ingredients. It may also arouse interests of researchers in the fields of medicine, pharmacy, agriculture, food, and so on.
Please find our paper at https://rdcu.be/evfJl for your reference.
References
- Wang, W., Xiao, H. & Zhong, J. Biosynthesis of a ganoderic acid in Saccharomyces cerevisiae by expressing a cytochrome P450 gene from Ganoderma lucidum. Biotechnol. Bioeng. 115, 1842–1854 (2018).
- Yuan, W. et al. Biosynthesis of mushroom-derived type II ganoderic acids by engineered yeast. Nat. Commun. 13, 7740 (2022).
Follow the Topic
-
Cell Discovery

This journal aims to provide an open access platform for scientists to publish their outstanding original works and publishes results of high significance and broad interest in all areas of molecular and cell biology.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in