Call for Paper: Integrating multi-omics to empower next-generation wheat breeding
Published in Genetics & Genomics, Plant Science, and Agricultural & Food Science

Aims and Scope
This topical collection centers on integrative omics in wheat, emphasizing how genomics, transcriptomics, epigenomics, proteomics, metabolomics, and phenomics, combined with systems biology and computational modeling, are reshaping wheat biology and breeding. The rapid growth of large-scale, high-quality omics datasets offers unprecedented power to elucidate gene function, regulatory networks, and the architecture of complex traits, thereby enabling precision strategies to improve yield, resilience, and grain quality. Against the backdrop of global food security, climate change, and sustainability imperatives, integrative omics provides a holistic view of genotype-phenotype-environment relationships. By connecting molecular layers to field performance, these approaches accelerate the discovery of causal variants and regulatory circuits, refine trait prediction, and inform the rational design of wheat ideotypes with enhanced productivity, stress tolerance, and nutritional value.
We seek innovative studies that integrate multiple omics in wheat or substantially enable such integration, offering a platform to share: original research that dissects complex traits and delivers breeding-ready targets, markers, models, or germplasm; FAIR-compliant data resources and databases (e.g., pangenomes, variation maps, single-cell/spatial omics); computational methods for cross-omics fusion, causal inference, network reconstruction, and genotype–environment modeling; big-data analyses, benchmarks, and interoperable pipelines; and concise reviews, perspectives, or methodologies that synthesize best practices and future directions.
Possible topics of interest include but are not limited to:
• Genomics, pangenomics, and structural variation analyses in wheat
• Transcriptomics, small RNA profiling, and regulatory network mapping
• Epigenomics, chromatin dynamics, and stress-responsive gene regulation
• Proteomics and metabolomics for functional characterization of key pathways
• Systems biology and network-based modeling of wheat traits
• Integrative multi-omics analyses to dissect traits in wheat
• High-throughput phenotyping and omics-assisted trait prediction
• Data-driven approaches linking omics datasets to wheat breeding pipelines
• Integration of big data and machine learning for trait prediction
All manuscripts will undergo rigorous peer review according to the journal’s standards. Accepted articles will be published in WheatOmics and included in this special collection, ensuring high visibility within the wheat and Triticeae research community.
Why submit to a Collection?
Submitting to a WheatOmics collection enhances the visibility and impact of your work. Curated by expert Guest Editors, collections provide a platform for high-quality research that is often more frequently cited and downloaded. Additionally, Article Processing Charges (APC) are sponsored for accepted manuscripts, eliminating publication fees for authors. This ensures your work freely reaches a global audience within the wheat and Triticeae research community, supported by our expert editorial team.
Who is involved?
Guest Editors:
- Shengwei Ma, PhD, Yazhouwan National Laboratory, China
- Weilong Guo, PhD, China Agricultural University, China
Schedule
Submission Open: 20th September 2025
Submission Deadline: 31st January 2026
Submission Procedure
All submissions will undergo a rigorous peer-review process to ensure the highest scientific quality. Manuscripts should follow the journal's guidelines for authors and be submitted through the online submission system.
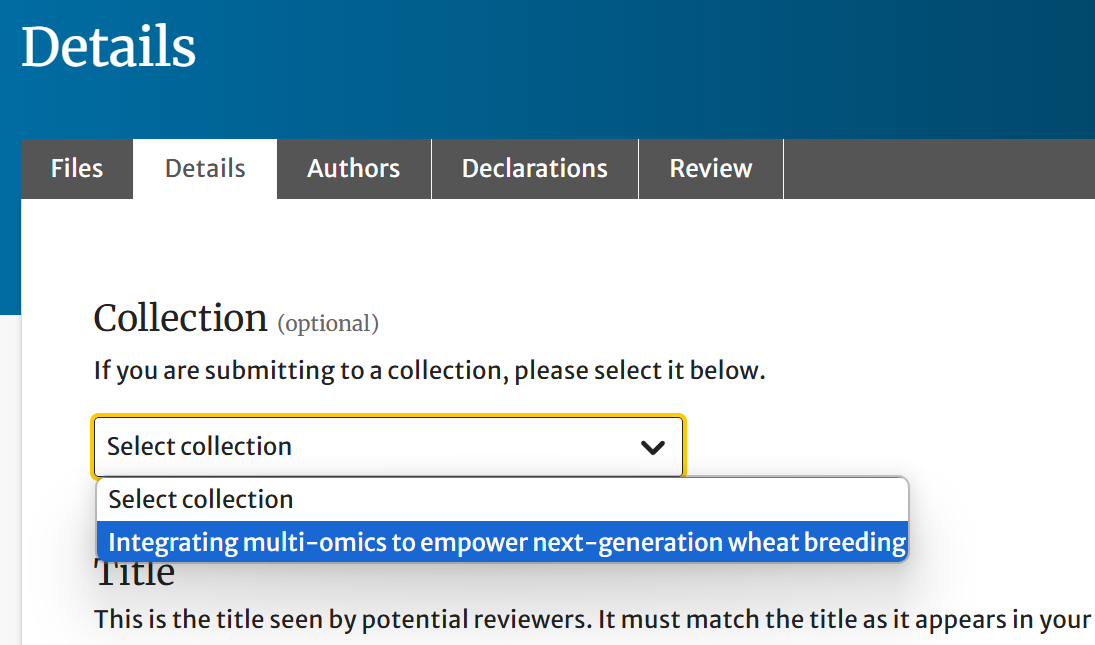
In the Details section, choose the appropriate collection from the dropdown/list provided.
Follow the Topic
-
WheatOmics

This journal is an open access international, peer-reviewed journal that publishes excellent, novel research on the broadest aspects of wheat and other Triticeae species.
Related Collections
With Collections, you can get published faster and increase your visibility.
Integrating multi-omics to empower next-generation wheat breeding
This topical collection centers on integrative omics in wheat, emphasizing how genomics, transcriptomics, epigenomics, proteomics, metabolomics, and phenomics, combined with systems biology and computational modeling, are reshaping wheat biology and breeding. The rapid growth of large-scale, high-quality omics datasets offers unprecedented power to elucidate gene function, regulatory networks, and the architecture of complex traits, thereby enabling precision strategies to improve yield, resilience, and grain quality. Against the backdrop of global food security, climate change, and sustainability imperatives, integrative omics provides a holistic view of genotype-phenotype-environment relationships. By connecting molecular layers to field performance, these approaches accelerate the discovery of causal variants and regulatory circuits, refine trait prediction, and inform the rational design of wheat ideotypes with enhanced productivity, stress tolerance, and nutritional value.
We seek innovative studies that integrate multiple omics in wheat or substantially enable such integration, offering a platform to share: original research that dissects complex traits and delivers breeding-ready targets, markers, models, or germplasm; FAIR-compliant data resources and databases (e.g., pangenomes, variation maps, single-cell/spatial omics); computational methods for cross-omics fusion, causal inference, network reconstruction, and genotype–environment modeling; big-data analyses, benchmarks, and interoperable pipelines; and concise reviews, perspectives, or methodologies that synthesize best practices and future directions.
Possible topics of interest include but are not limited to:
• Genomics, pangenomics, and structural variation analyses in wheat
• Transcriptomics, small RNA profiling, and regulatory network mapping
• Epigenomics, chromatin dynamics, and stress-responsive gene regulation
• Proteomics and metabolomics for functional characterization of key pathways
• Systems biology and network-based modeling of wheat traits
• Integrative multi-omics analyses to dissect traits in wheat
• High-throughput phenotyping and omics-assisted trait prediction
• Data-driven approaches linking omics datasets to wheat breeding pipelines
• Integration of big data and machine learning for trait prediction
Publishing Model: Open Access
Deadline: Sep 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in