Century-Old Mystery Solved: How a Single Somatic Cell Becomes Totipotent and Regenerates into a Whole Plant
Published in Genetics & Genomics, Plant Science, and Agricultural & Food Science
WheatOmics, an international open access journal dedicated to advancing wheat research, with APC supported by Shandong Agricultural University sponsorship, is delighted to share a landmark discovery from our Editor-in-Chief. How does a single plant cell restart its life and develop into an entire plant? This century-old question has fascinated plant scientists for generations and was recognized by Science in 2005 as one of the 125 most important unanswered problems in biology.
Now, a research team led by our Editor-in-Chief Professor Zhang Xiansheng and Professor Su Yinghua at Shandong Agricultural University has finally cracked the mystery. Their landmark study, published in renowned academic journal Cell (September 16, 2025). They fully revealed the molecular mechanism that allows a single somatic cell to reprogram into a totipotent state and regenerate into a whole plant. This breakthrough not only solves the long-standing puzzle of plant cell totipotency but also establishes a new theoretical foundation for genetic improvement, rapid crop propagation, and efficient plant regeneration.
How does a cell "restart" its life program?
In 1902, the concept of "plant cell totipotency" was proposed; stating that plant cells can dedifferentiate into totipotent stem cells, similar to fertilized eggs, and subsequently develop into complete plants. This remarkable ability occurs widely across the plant kingdom, including in crops and woody plants, yet the molecular mechanisms driving totipotency have remained largely unexplained for over a century.

Professor Zhang Xiansheng, the corresponding author of the study, explained that plant cells exhibit greater developmental plasticity than animal cells. Under certain conditions, they can develop into embryos without fertilization, a process known as somatic embryogenesis. Plant cells also possess a remarkable ability to regenerate: after undergoing reprogramming, somatic cells from a leaf can revert to a stem-cell-like state and then enter the somatic embryogenesis stage, eventually developing into a complete plant. This regenerative ability has significant practical applications in agriculture, including rapid propagation, biotechnology-based breeding, and virus-free plant culture. Yet the core mechanism by which ordinary somatic cells are reprogrammed into totipotent embryos has remained unrevealed.

"Just like a leaf, which should remain a leaf forever, can ‘transform’ into a whole new plant, how does this reversal of fate occur?" asked Su Yinghua, one of the corresponding authors and a Changjiang Scholar Distinguished Professor at Shandong Agricultural University, explaining the motivation behind the research. Under the guidance of Professor Zhang Xiansheng, the team began studying Arabidopsis thaliana as a model in 2005, embarking on what would become a 20-year scientific research journey.
From Accidental Discovery to Perfect System
In the early stages of the research, the biggest challenge was establishing an experimental system that would allow single somatic cells to directly develop into embryos. In 2009, the team discovered for the first time in Arabidopsis that the accumulation of high levels of auxin acts as a “switch” to activate cell totipotency, with the findings published in the Plant Journal.
A turning point came in 2011 with an unexpected observation. An induction factor caused embryonic structures to grow directly on the surface of seedling leaves. This somatic embryo actually originated directly from a single cell on the surface of the leaf. This accidental phenomenon made Professor Su Yinghua realize that mature leaf cells can be directly induced into embryos without the need for callus tissue transition. To reproduce this phenomenon reliably, Professor Su Yinghua and his team conducted repeated verifications, “as if restarting the experiment,” eventually establishing a stable system for inducing somatic embryogenesis from a single cell.

The next challenge was identifying a molecular marker for totipotent stem cells. After numerous verification experiments, the team successfully discovered a fluorescent marker that glows only in totipotent stem cells, providing a powerful tool for tracking and studying these cells.
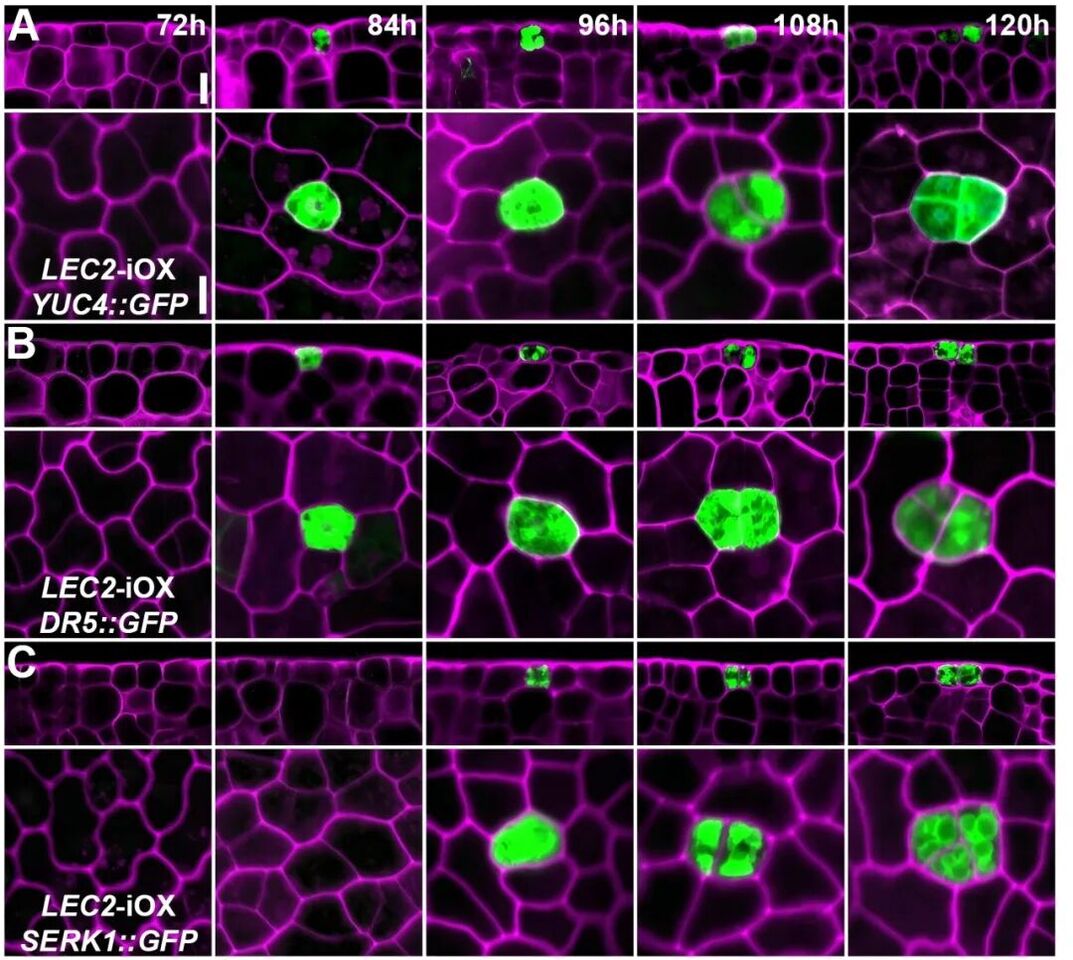
 Somatic embryos are derived from a single totipotent stem cell
Somatic embryos are derived from a single totipotent stem cell
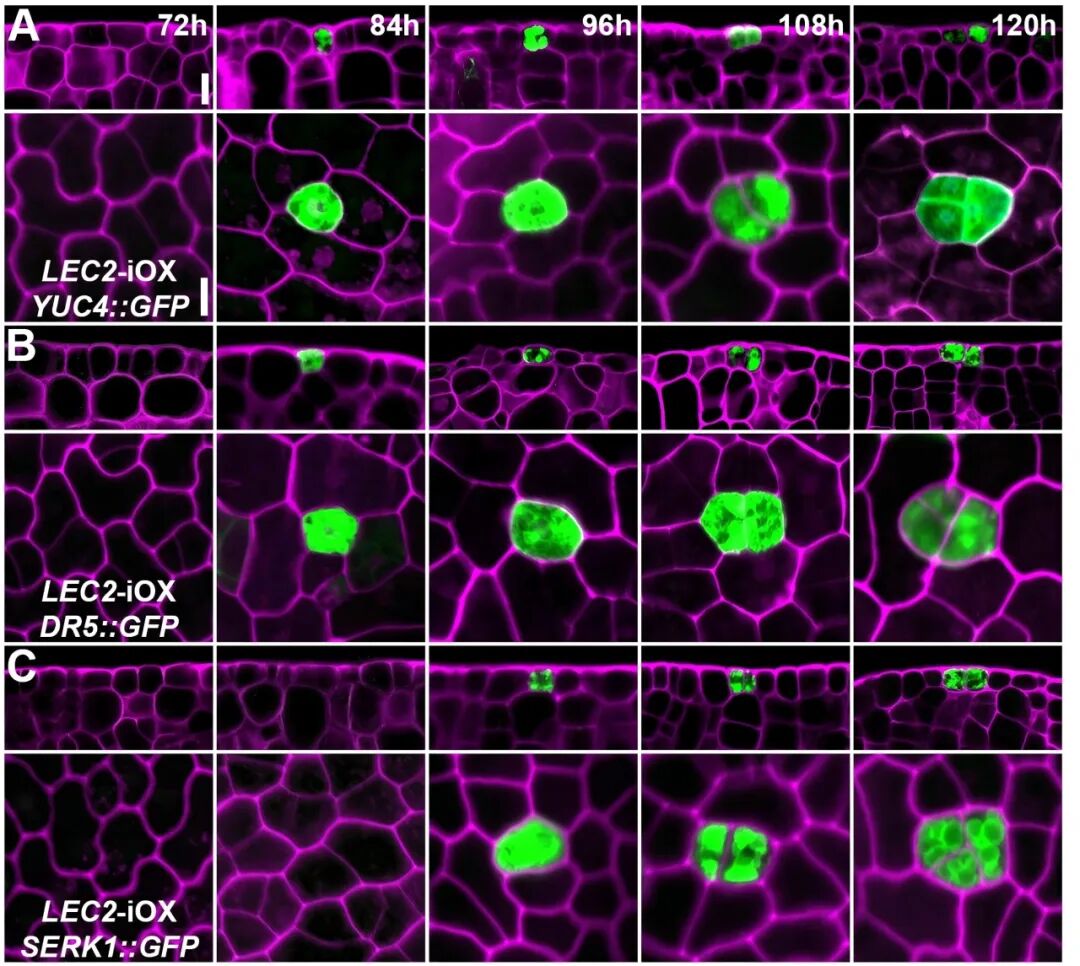 Auxin accumulation in stomatal precursor cells transforms them into totipotent stem cells.
Auxin accumulation in stomatal precursor cells transforms them into totipotent stem cells.
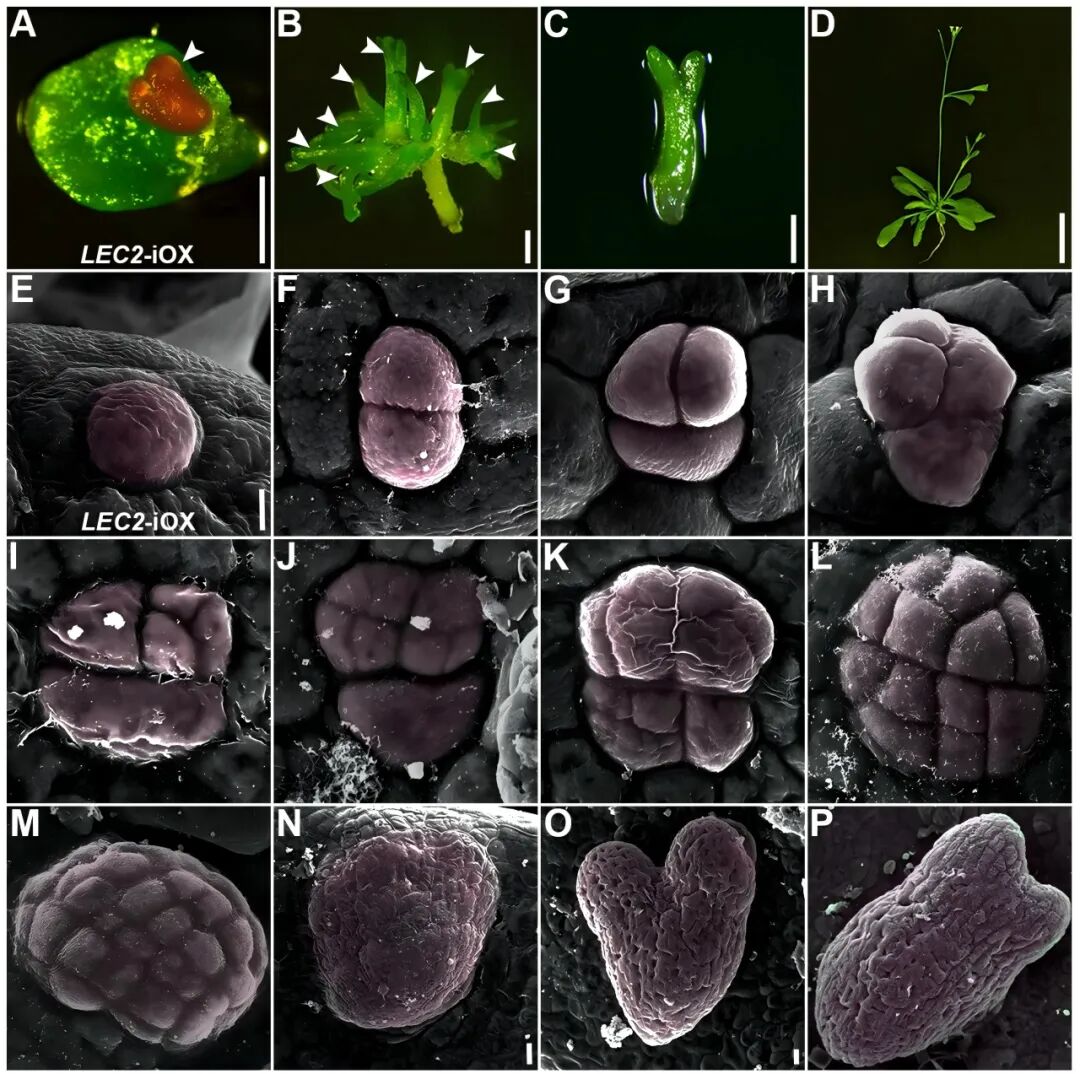
The establishment of a stable induction system and the identification of totipotent stem cell markers paved the way for deeper exploration. Using scanning electron microscopy and laser confocal microscopy, the team captured, for the first time, the entire process of single plant cell division: from one cell to two, and then, in a distinctive “groups of three” pattern, to form a 12-cell embryoid. This visualization provides clear evidence of the single-cell origin of plant cell totipotency, effectively resolving a long-standing scientific question.
How do two key genes "trigger" totipotency?
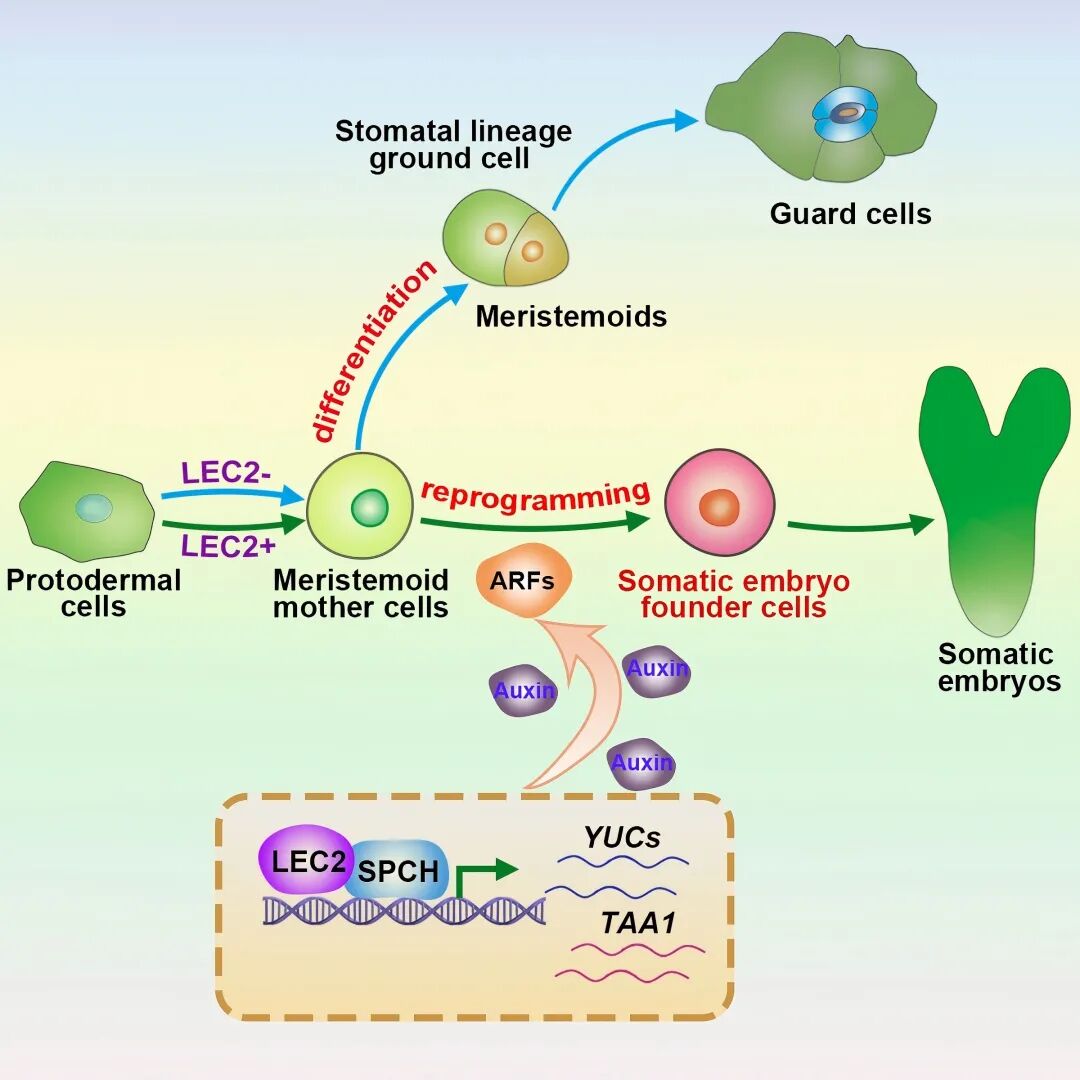
Through in-depth research, the team identified the key drivers of cellular totipotency: the gene SPCH, specific to leaf stomatal precursor cells, and the artificially induced high-expression gene LEC2. These two genes work synergistically to form a “molecular switch.” Professor Zhang Xiansheng described it elegantly: “It is like turning a lock that requires two keys; neither works without the other.”
Associate Professor Tang Liping, the paper’s first author, explained that somatic embryos originate from a single totipotent stem cell. This “precursor cell,” originally destined to develop into a stoma, activates the auxin synthesis pathway through the combined action of LEC2 and SPCH. This leads to a localized accumulation of auxin, causing the precursor cell to deviate from its stomatal development trajectory and become a totipotent stem cell capable of generating new life.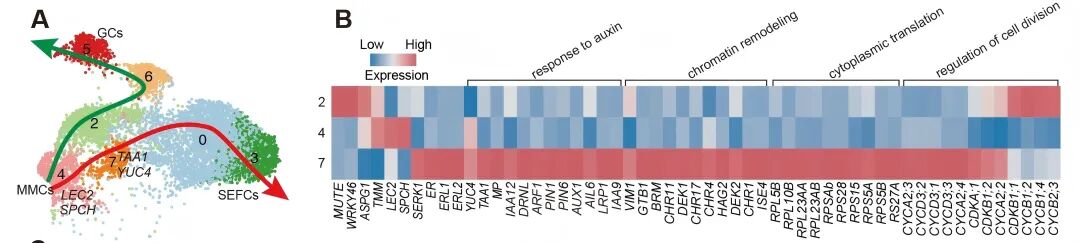
Two Developmental Paths of Stomatal Precursor Cells
Using cutting-edge technologies such as single-cell sequencing, microdissection transcriptome sequencing, and in vivo imaging, the team fully documented the process of cell fate remodeling. They identified a critical bifurcation point: one pathway allows stomatal precursor cells to continue differentiating into stomata, while the other, driven by the accumulation of endogenous auxin, reprograms individual somatic cells into totipotent stem cells that enter the embryogenesis pathway.
 A schematic diagram illustrates the two developmental pathways of stomatal precursor cells.
A schematic diagram illustrates the two developmental pathways of stomatal precursor cells.
Researchers named this critical transitional state the “GMC-auxin” intermediate. In this state, cells undergo extensive chromatin remodeling, gradually activating many previously silent genes. This leads to a divergence in cell fate trajectories, paving the way for the establishment of totipotency. Further experiments showed that blocking endogenous auxin synthesis completely prevents reprogramming and somatic embryo formation. Moreover, adding exogenous hormones could not substitute for the auxin produced by the cells themselves, highlighting that only cell-autonomous auxin signals can initiate totipotency.
More importantly, this study is the first to comprehensively reveal the molecular mechanisms by which single plant somatic cells reprogram into totipotent stem cells and regenerate entire plants. In the GMC-auxin intermediate, multiple transcription factors form a tightly coupled regulatory network, activating downstream embryogenesis programs.
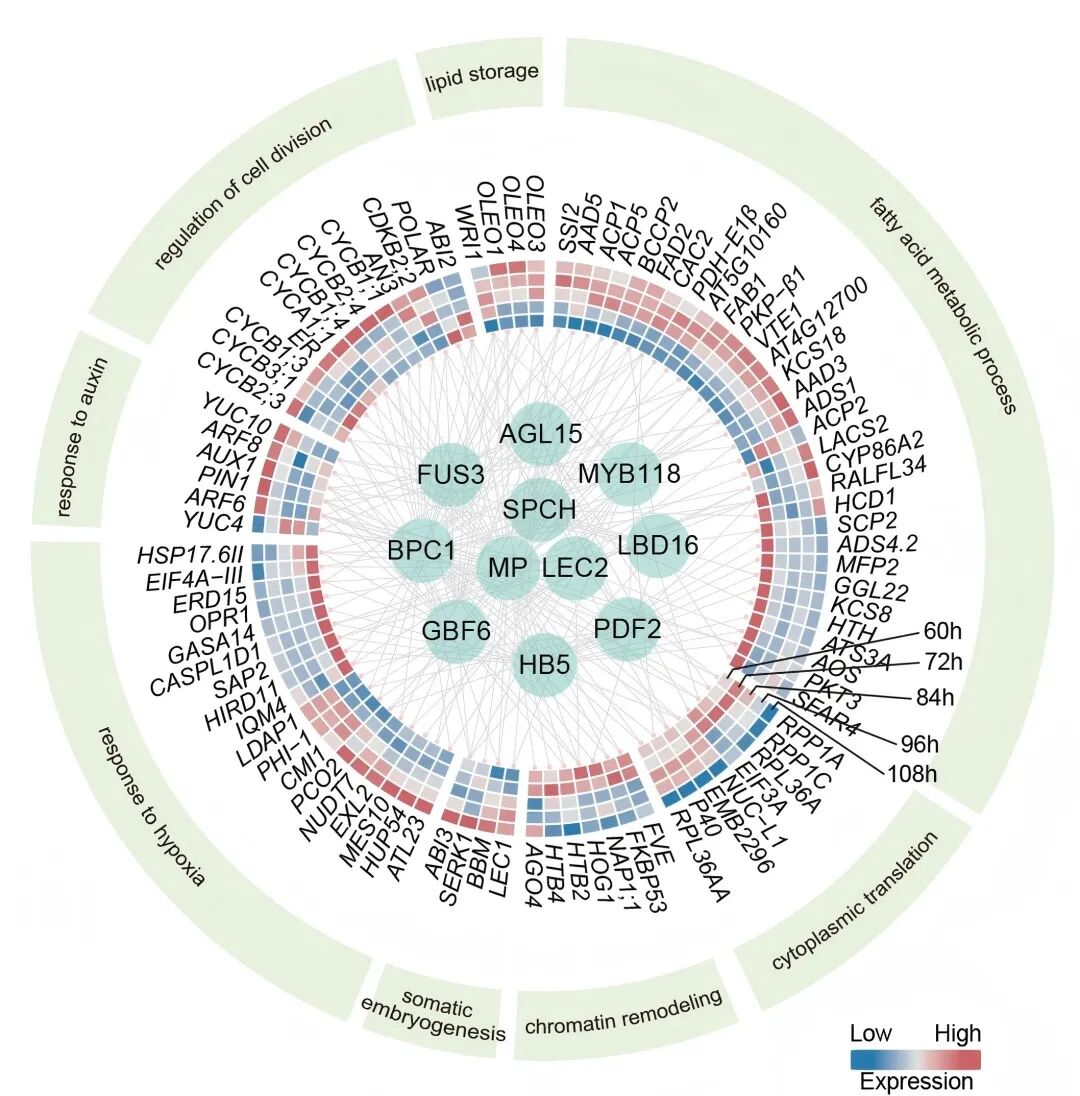
Molecular Regulatory Network for Reprogramming Plant Somatic Cells into Totipotent Stem Cells
Reviewers of the journal Cell described the identification of the GMC-auxin intermediate as an “exciting breakthrough,” providing the first clear molecular pathway for transforming stomatal precursor cells into totipotent stem cells. This highly original study offers strong scientific insight into cell fate changes and the regenerative potential of plant somatic cells.
Accelerating Smart Breeding from the Laboratory to the Field
This study not only advances our understanding of the fundamental principles of plant cell development but also provides new strategies and technical tools for precisely controlling plant regeneration and improving crop traits.
Experiments using this system are now underway in major crops, including wheat, corn, and soybeans. “In the future, precise control of cell totipotency could enable the rapid cloning of superior crop varieties, dramatically shortening breeding cycles and supporting precision design breeding,” said Professor Zhang Xiansheng. “It will also provide new opportunities for the efficient conservation of rare plant germplasm and for advancing plant synthetic biology.”

Meet the paper's lead authors are: From left: Postdoctoral Fellow Zhai Liming, Professor Su Yinghua, Associate Professor Zhang Wenjie, Professor Zhang Xiansheng, Dr. Gao Yue, Associate Professor Tang Liping, and Professor Tian Xin.
The study was led by Professors Zhang Xiansheng and Su Yinghua at Shandong Agricultural University, with co-corresponding authors from Radboud University, the BGI-Research Institute of Life Sciences, and other collaborators.
We congratulate our Editor-in-Chief Professor Zhang Xiansheng and Editorial Board member Professor Su Yinghua, along with their research team, on this remarkable achievement. Their groundbreaking study, published in Cell, not only solves a century-old puzzle in plant biology but also opens new avenues for precision crop breeding and plant regeneration.
Read the full article in Cell: https://doi.org/10.1016/j.cell.2025.08.031
Follow the Topic
-
WheatOmics

This journal is an open access international, peer-reviewed journal that publishes excellent, novel research on the broadest aspects of wheat and other Triticeae species.
Related Collections
With Collections, you can get published faster and increase your visibility.
Integrating multi-omics to empower next-generation wheat breeding
This topical collection centers on integrative omics in wheat, emphasizing how genomics, transcriptomics, epigenomics, proteomics, metabolomics, and phenomics, combined with systems biology and computational modeling, are reshaping wheat biology and breeding. The rapid growth of large-scale, high-quality omics datasets offers unprecedented power to elucidate gene function, regulatory networks, and the architecture of complex traits, thereby enabling precision strategies to improve yield, resilience, and grain quality. Against the backdrop of global food security, climate change, and sustainability imperatives, integrative omics provides a holistic view of genotype-phenotype-environment relationships. By connecting molecular layers to field performance, these approaches accelerate the discovery of causal variants and regulatory circuits, refine trait prediction, and inform the rational design of wheat ideotypes with enhanced productivity, stress tolerance, and nutritional value.
We seek innovative studies that integrate multiple omics in wheat or substantially enable such integration, offering a platform to share: original research that dissects complex traits and delivers breeding-ready targets, markers, models, or germplasm; FAIR-compliant data resources and databases (e.g., pangenomes, variation maps, single-cell/spatial omics); computational methods for cross-omics fusion, causal inference, network reconstruction, and genotype–environment modeling; big-data analyses, benchmarks, and interoperable pipelines; and concise reviews, perspectives, or methodologies that synthesize best practices and future directions.
Possible topics of interest include but are not limited to:
• Genomics, pangenomics, and structural variation analyses in wheat
• Transcriptomics, small RNA profiling, and regulatory network mapping
• Epigenomics, chromatin dynamics, and stress-responsive gene regulation
• Proteomics and metabolomics for functional characterization of key pathways
• Systems biology and network-based modeling of wheat traits
• Integrative multi-omics analyses to dissect traits in wheat
• High-throughput phenotyping and omics-assisted trait prediction
• Data-driven approaches linking omics datasets to wheat breeding pipelines
• Integration of big data and machine learning for trait prediction
Publishing Model: Open Access
Deadline: Sep 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in