Catching RNA in the Act of Folding
Published in Cell & Molecular Biology
RNA plays a fundamental role in life by performing many vital functions including regulation of genetic material, catalysis of biological reactions, recognition of biomolecules, and protein synthesis. But to perform these functions, RNA often has to adopt specific three-dimensional (abbreviated as 3D) structure.
For a long period of time, RNA’s ability to acquire stable 3D structures was not appreciated, possibly due to a dogma that considered RNA to be too flexible. But this vision is changing now and an exponentially increasing number of RNA structures is becoming available. Still, it is true that RNA dynamics are crucial to understand RNA structural-to-functional relationships, and yet very little experimental evidence is available until now to describe RNA dynamics, especially at near-atomic resolution. A particularly poorly-understood step in RNA structural biology is the mechanism by which RNA dynamically adopts well defined 3D structures, i.e. the process of RNA folding.
In this context, our study, “Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy” published in Nature Communications, is a timely and unique contribution, because it provides a molecular movie of the dynamics of a large RNA in the process of folding.
Our study focuses on a catalytic RNA called the self-splicing group II intron – a molecular machine capable of self-excision from RNA transcripts. Group II introns are present in bacteria, archaea and eukaryotic organelles, like chloroplast and mitochondria from plant and fungi. They interrupt essential genes, thus defects in group II intron splicing can be lethal for the host organisms. Moreover, the structure of the active site and the catalytic process of splicing is not only conserved across group II introns, but more broadly also in other splicing machineries like the human nuclear spliceosome.
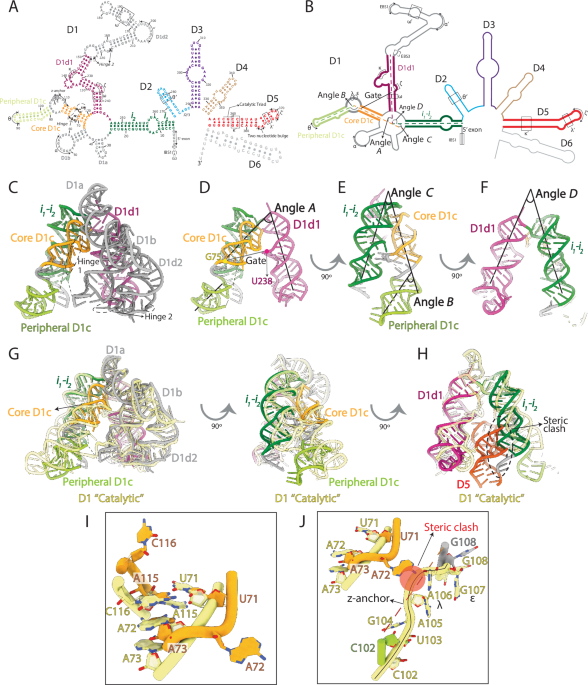
About 15 years ago, the first 3D structure of a group II intron was determined. The structure revealed that group II introns are organized into five domains (called D1 to D5), with D1 possessing an inverted V shape structure, and acting as a scaffold for peripheral domains D2, D3, and D4. The last domain, D5, docks in the core of D1 forming the splicing active site. Those earlier studies also showed that, before assembling with the other domains, D1 temporarily adopts a closed structure which is incapable of binding to D5. This suggested that D1 is a dynamic scaffold and – thanks to the effect of D2, D3, and D4 - needs to open its core to bind D5 and form a functional splicing active site.
To unravel this domain assembly process, we leveraged the use of electron cryo-microscopy (abbreviated as cryo-EM) and developed a protocol to enable visualization of RNA and RNA dynamics. Indeed, at the beginning of our work there were no available cryo-EM structures of RNA alone, but only in the context of protein-RNA complexes. The Marcia group thus established a new cryo-EM workflow, to flash freeze our target RNA in thin ice and visualize its 3D structure in three different folding states at near-atomic resolution, namely the D1-2, the D1-3 and the D1-4 states. In these states, we could unexpectedly capture novel conformations of D1, sampling dynamic movements of its conserved helices. Molecular simulations on these large systems – carried out with the help of collaborators from the De Vivo group at IIT – helped depicting the exact energy landscape of group II introns. To reveal near-atomic details of the conformational movements of D1, though, conventional cryo-EM image processing was not enough. Here, a serendipitous encounter made the difference. During the 2023 CCP-EM symposium, Shekhar and Thomas met, and that interaction seeded a novel collaboration between the Marcia and the Topf group. As a result of that collaboration, we could improve our image analysis and reveal the role of specific residues in regulating group II intron dynamics. These results were crucial to stimulate the design of biochemical and enzymatic assays, aimed at engineering group II introns, perturb their folding and look at resulting effects on catalysis.
The integrated combination of RNA biochemistry, RNA cryo-EM imaging, advanced image processing and molecular simulations lead us to the formulation of a molecular mechanism for group II intron folding that is likely applicable to many other RNA molecules. The implications of the study are manifold. For instance, we could identify new potential target sites for antibiotic agents, and we produced six new experimental high-resolution RNA structures that will inform the development of machine learning algorithms for RNA structure prediction.
Jadhav, S., Maiorca, M., Manigrasso, J. et al. Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy. Nat Commun 16, 10195 (2025).
https://www.nature.com/articles/s41467-025-65502-8
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in