“Effective interactions” shape the evolution of drug resistance (and much more)
Published in Ecology & Evolution, Microbiology, and Protocols & Methods
Recently, I had the pleasure of participating (virtually) in a workshop at National Human Genome Research Institute (NHGRI) focused on the genetic architecture of complex traits. It featured many of the world’s leading population genetics, statisticians, and many others. One of the themes from the discussion involved ways to carefully consider the role of the environment in shaping complex phenotypes (e.g., disease, behavior).
In conversations like these, I often feel like “the environment” becomes a black box where we store much of what we don’t know or can’t measure in genetics--what accounts for our inability to detect neat and genomic signals for a complex trait? "The environment!" (whatever that means)
In a new study published in Nature Communications, me and two close colleagues asked an important question about this black box: “What if we could study the role of the environment at a smaller scale? Could we come up with a new and interesting way to measure how the environment shapes evolution? And could we relate this to a practical problem, like disease?”
A few concepts were central to our thinking about this problem.
-
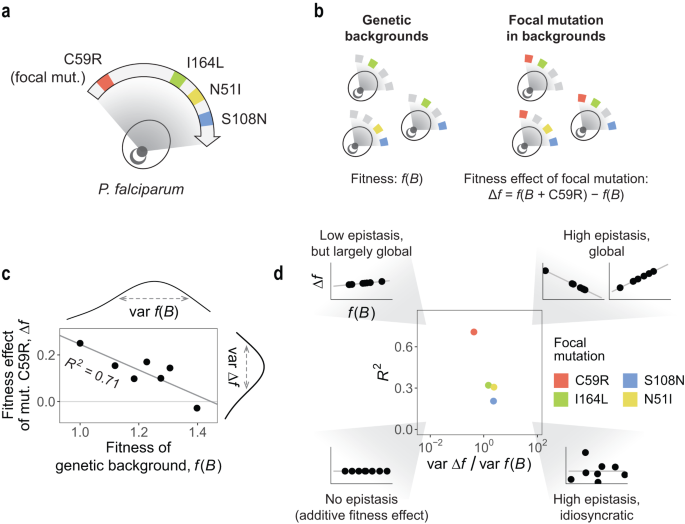
One is the concept of the “adaptive or fitness landscape.” It’s an analogy for genotype-phenotype maps that analogizes evolution as moving “uphill” or “down valleys.” In this analogy, spaces on the ground are genotypes, and the heights correspond to some value related to “fitness.”
-
The second is the concept of “epistasis.” While I wouldn’t quote call it “controversial,” it is a provocative idea (despite being over a century old). It posits that parcels of genetic information (genes, mutations) can interact in a nonlinear or non-additive fashion, which complicates our ability to predict how Gene variant [A] or [B] will affect a phenotype. It turns out that the effects of a gene or mutation are influenced by the existence of other genes or mutations. (I urge those who want to learn more to check out an article I wrote on epistasis for Quanta Magazine), and a fun analogy blog that me and a colleague wrote about epistasis at few years back).
-
Last is an idea called “global epistasis.” It was developed relatively recently, and describes a phenomenon whereby the effects of mutations might be nonlinear/non-additive (as in epistasis), but still follow a tractable pattern–they depend on the fitness of the background organisms/genome to which they are being added. That is, there is an underlying rhyme and reason (that can be described mathematically) for how the unpredictability of epistasis manifests. It has been observed in a number of biological settings, and is exciting because it tames the frustration with the idea that effects of mutations are always dependent on a specific genetic background.
Our new study was led by Juan Diaz-Colunga, a postdoctoral associate currently at the Institute of Functional Biology and Genomics IBFG-CSIC, University of Salamanca (formerly at Yale and the Spanish National Center for Biotechnology). In it, we propose a framework for measuring how drug environments influence global epistasis in a fitness landscape corresponding to mutations in a malaria parasite (Plasmodium falciparum) enzyme that is associated with resistance to antimalarial drugs (pyrimethamine and cycloguanil).
How did this project come together? The story begins when me and Alvaro Sanchez co-taught an upper-division evolutionary theory course while faculty colleagues at Yale University during the spring semester of 2021.
I had been a long-time admirer of Sanchez’s work, and this course presented the perfect opportunity for us to meet regularly and discuss our common interests. Sanchez had already established himself as one of the world’s best thinkers on topics related to how we can take population genetics principles and apply them to thinking about interactions in the microbiota. For our evolutionary theory course, we challenged ourselves to learn and teach cutting-edge topics in theoretical population genetics.
We used a plethora of resources, including the classic John Gillespie textbook that contained mathematical formalisms. And among the key topics of this course was epistasis. During this time, Sanchez and members of his research group had begun to publish what would be some groundbreaking work on the application of the fitness landscape analogy to the study of microbial communities. And Sanchez (a physicist by training) had started to develop an interest in global epistasis, and ways to apply this to microbial communities.
Before Sanchez and his group moved to the Spanish National Center for Biotechnology CNB-CSIC in 2022, I had the pleasure of meeting Dr. Diaz-Colunga. He was also a physicist, and had established a reputation for his work in developing physics-inspired theory for studying evolutionary theory. He had grand ideas for how global epistasis applied to fitness landscapes. Before moving to Spain, he spent time as a visiting postdoctoral associate in my research group.
Sanchez, Diaz-Colunga and I exchanged thoughts on the topic. We developed questions about applying the global epistasis framework to how we think about the role of the environment in shaping molecular evolution. We explored the literature, and there were only passing mentions of the topic, little or no direct explorations.
Using a dataset that was analyzed in prior studies of fitness landscapes in antimalarial drug resistance, Diaz Colunga applied the global epistasis framework to accomplish something very important: rigorously describe the role of the environment in crafting interactions between mutations, using an analytical, global epistasis framework. The analysis turned up some interesting findings.
Firstly, we analyzed a fitness landscape of intrinsic biological significance (the four mutations confering resistance to antifolate drugs in P. falciparum). This has value because it pushes the "global epistasis framework" (a relatively new idea) beyond model organisms & into the disease world. Until now, its applications have largely been relegated to model systems or theoretical settings. We believe that this is one of the first explicit disease-focused applications of global epistasis (one prior study focusing SARS-CoV-2).
Secondly, the findings demystify the role of the environment. We show that environmental effects on global epistasis can be readily traced back to specific gene-by-environment and gene-by-gene-by-environment interactions, and the contribution of each mechanism can be readily quantified under this framework.
Lastly (and to get a tad more technical): we think there is an important conceptual advance in the way we define this "distribution of effective interactions." Classical examinations of mutation effects can look at the "raw" distributions of fitness effects or distributions of epistatic effects, and that of course is informative. Alternatively, looking at the "distribution of effective interactions" (where we define an "effective interaction" between as the average epistasis relative to the average fitness effect, what we show in the main manuscript Box 1) allows us to express the overall strength of epistasis, the degree of global epistasis, and the shape of global epistasis simply from the (weighted) moments of the distribution.
Here’s what we mean: when determining how important epistasis is, we often measure it at face value. But an “effective interaction” focuses on the strength of epistatic effects relative to the average effects of mutations. That is, quantifying the effect of interacting mutations or genes is important. But what about their effect relative to the average standard effect? This “effective interactions” calculation puts things into perspective.
Broadly, the findings have implications for both evolutionary theory and biomedicine. For theoreticians, this paper makes what I believe is one of the most direct examinations of how the environment shapes global epistasis. Why is this relevant? Let’s go back to my experience at the NHGRI meeting.
At this meeting, some of us struggled to find language for how to describe (let alone measure) how environments might influence complex biological traits. While our study doesn’t speak to anything related to human genomics, our work does offer an important first step in the quest to carefully disentangle how the world we are in (the environment) influences how genes function.
In the biomedical context, our work can help to resolve the rules governing how pathogen resistance to antimicrobial drugs happens. In the long run, this may help us extract fundamental rules to how resistance happens, which can help us design new therapies that take advantage of knowledge of the rules, to be more effective.
But as substantial as these formal “scientific” lessons are, they aren’t the true spirit behind this paper. In technical terms, this work is an example of the delightful nonlinear interactions that can occur between objects or entities. Sometimes these entities are mutations. Sometimes the objects are microbes. And as we have demonstrated, these interactions often have fundamental rules that guide them, as in global epistasis. But what about when the agents interacting are....scientists?
When it comes to people, the only rule governing the success of “effective interactions” is that they are fun. And this might be the most important theoretical axiom of all.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in