Fast and effective molecular property prediction with transferability map
Published in Chemistry
Challenge
Molecular property prediction has been widely considered as one of the most critical tasks in computational drug and materials discovery, as many methods rely on predicted molecular properties to evaluate, select and generate molecules. With the development of artificial intelligence (AI), effective transfer learning for molecular property prediction exhibits a great advantage in addressing insufficient labeled molecules. However, many existing methods are still challenged by their inability to effectively account for the quantitative relationship between source and target properties, risking negative transfer, or necessitate intensive training on target tasks. Thus, a fast and effective method for quantifying the suitability of the source property for the target property prior to training on the target task is required.
Approach
To address the challenge of transfer learning for molecular property prediction, we propose a simple, fast, and effective Principal Gradient-based Measurement (PGM) to quantify the transferability from the source property to the target property (Figure 1). First, inspired by the predictive role of gradients in capturing intrinsic task-related characteristics for model optimization, we design a restart scheme to calculate a principal gradient in an optimization-free manner. The distance between the principal gradient obtained from model training on the source dataset and that derived from the target dataset indicates transferability. Second, we build a quantitative transferability map by performing PGM on various molecular property prediction datasets to show the inter-property correlations in property space distribution. The map is extensible and can be a reference standard for transfer learning in molecular property prediction, even when applied to a few target samples. Third, through the map, we can capture and transfer the most desirable source dataset for the given target dataset, so as to promote performance on the target task and avoid negative transfer.
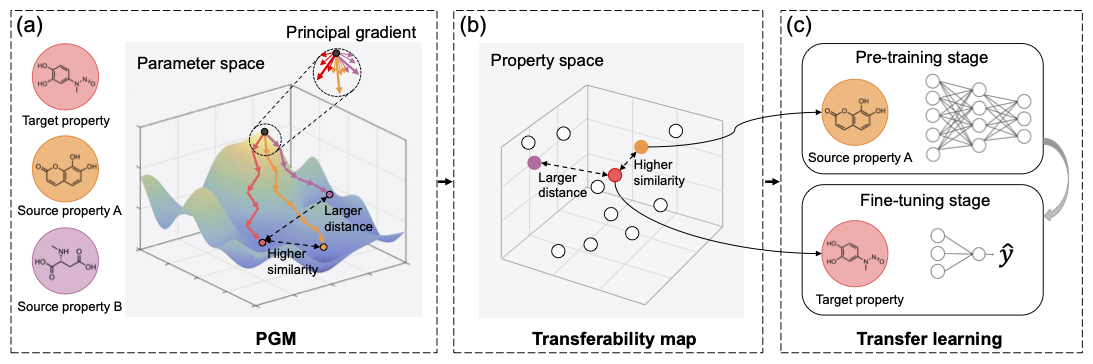
Figure 1. Illustration of Principal Gradient-based Measurement (PGM) for guiding transfer learning in molecular property prediction.
Results
We evaluate PGM thoroughly on 12 benchmark datasets from MoleculeNet with various molecular property prediction tasks. We build a quantitative transferability map to intuitively observe the task-relatedness between these molecular property prediction datasets. Then we perform a transferability map-guided cross-task transfer learning strategy. Specifically, each of the 12 datasets is used as the target dataset, while the remaining 11 datasets are employed as source datasets, as described below. Initially, the model is trained on each source dataset to obtain pre-trained models. Subsequently, each of these pre-trained models is fine-tuned on the target dataset. As depicted in Figure 2, a significant correlation between the predicted transferability and the transfer learning performance across various tasks can be observed.
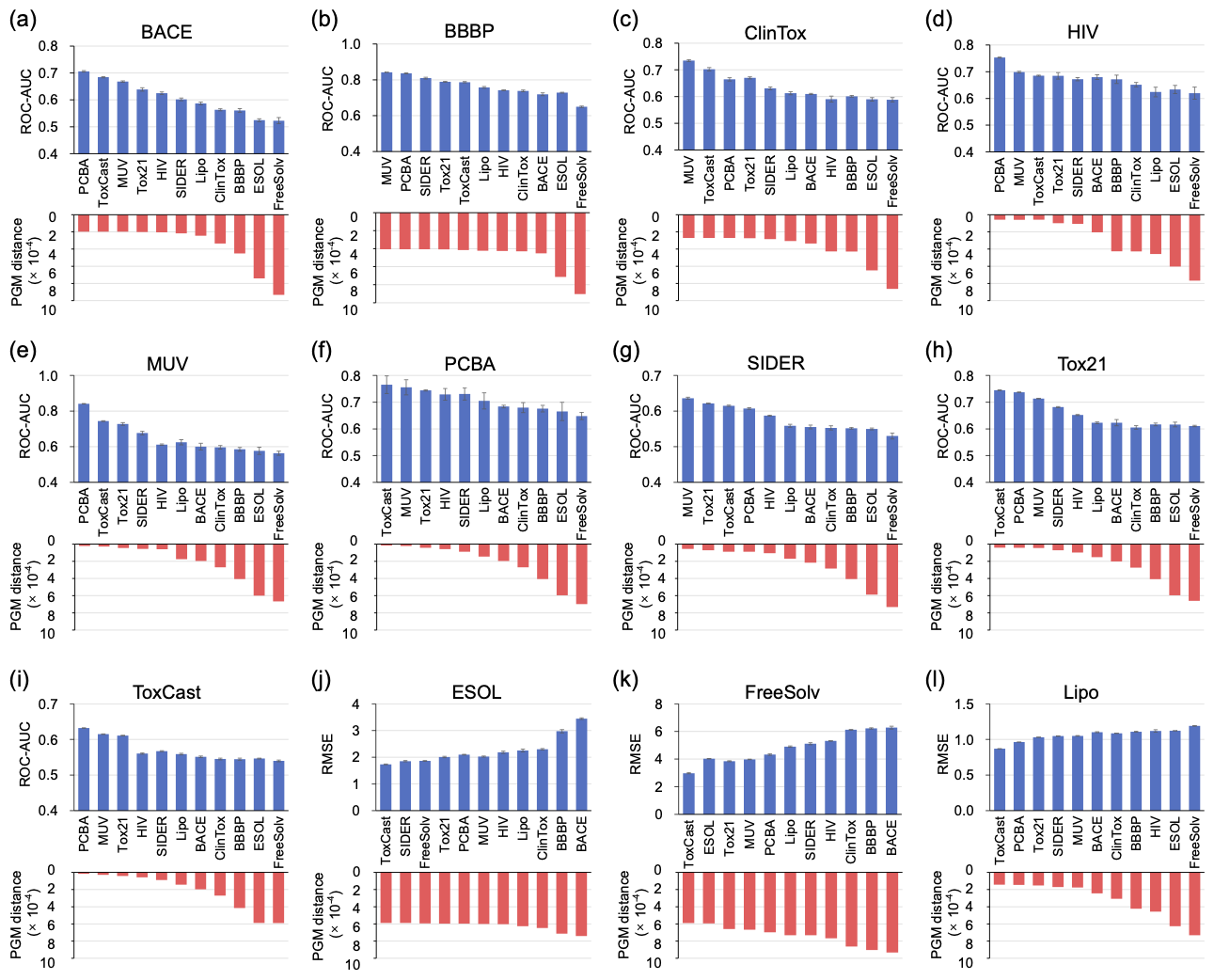
Figure 2. Comparison of the PGM distance and the transfer performance on the 12 target datasets.
Furthermore, we expand the transferability map's application from the above properties to subtasks within them. We also investigate the effectiveness of each module in PGM by conducting ablation studies focusing on three key areas: the computation efficiency of PGM, its performance relative to the size of the target dataset, and its behavior in relation to varying dataset sizes across different tasks. As resulted, the proposed approach can serve as fast and effective guidance to enhance the transfer performance of molecular property prediction.
Highlights
We propose a method to support transferability quantification for molecular property prediction datasets. Specifically, we design a principal gradient to approximate model optimization, which performs on source and target datasets to realize transferability measure between datasets. Furthermore, we build a transferability map based on PGM to access task-relatedness prior to applying transfer learning. Both theoretical and empirical studies demonstrate that PGM strongly correlates with the transfer performance of molecular property prediction, making it a quantified transferability measure for source dataset selection. This work contributes to more efficient discovery of drugs, materials, and catalysts by offering a task-relatedness quantification prior to transfer learning and understanding the relationship between chemical properties.
For more detail on the experiments and results, please read our paper:
https://www.nature.com/articles/s42004-024-01169-4
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Advances in Asymmetric Catalysis for Organic Chemistry
Publishing Model: Open Access
Deadline: Mar 31, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in