Genome wide association study unveils the genetic basis of Orobanche crenata resistance in pea
Published in Genetics & Genomics, Plant Science, and Agricultural & Food Science
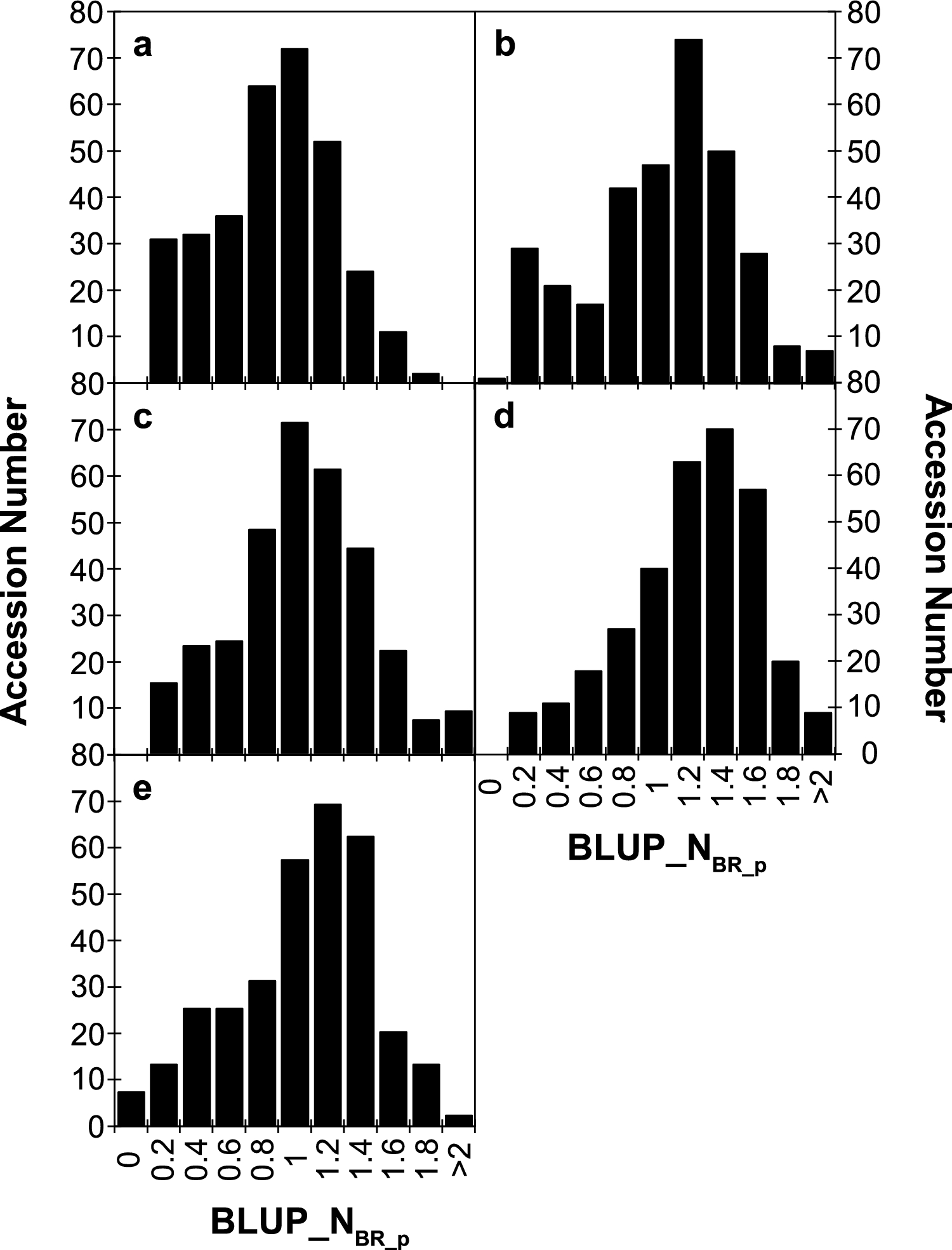
This study presents the first genome-wide association study (GWAS) for O. crenata resistance in pea. We genotyped 324 diverse pea accessions with DArT markers and phenotyped over four field seasons. Substantial phenotypic variation and environmental influence were observed, with wild Pisum fulvum and P. sativum subsp. elatius emerging as key resistance sources. GWAS identified 73 significant marker–trait associations, with Chromosome 5 as a major resistance hotspot. Several loci co-localized with previously reported QTLs, reinforcing their reliability. Interestingly, 24 candidate genes were linked to resistance, including those involved in vesicle trafficking, transcriptional regulation, and defense signaling (notably LRR receptor-like kinases). These findings deliver new genetic insights and molecular markers for breeding broomrape resistant pea cultivars. It also provide a foundation for functional candidate gene studies and advancing sustainable pea improvement for food security.
Note: Read more about this research in our recent article published in Theoretical and Applied Genetics.
https://link.springer.com/article/10.1007/s00122-025-05051-2
Follow the Topic
-
Theoretical and Applied Genetics

Theoretical and Applied Genetics focuses on modern plant genetics, genomics and biotechnology with significant impact on plant breeding.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in