How does human disturbance impact plant-associated pathogens, and why does it matter?
Published in Microbiology

Evidence is accumulating that pathogens, although defined by their harmful effects on individual host plants, are essential to maintaining plant community diversity. Through these effects, pathogens may mediate plant community responses to climate change and disturbance. Nonetheless, little is known about how pathogen communities, especially those directly associating with plants in roots, respond to climate and human land use. Therefore, we sequenced root pathogens across a Midwestern US precipitation and temperature gradient, comparing undisturbed and human disturbed grasslands. We specifically looked at changes in two groups of root-associated plant pathogens: fungal pathogens and oomycetes (an important group of pathogens known mostly for being agricultural pests).
I first started working on this project four years ago, at the start of my PhD. Through this project, I developed my molecular toolset, alongside my long-term undergraduate assistant, lab technician and mentee, Josh Schemanski (Fig. 1). We worked through amplifying DNA (for the first time), cleaning the samples, adapting a bioinformatical pipeline, answering our questions, and going through the publication process. It is also through this project that I really got to know several post-docs and professors at the Kansas Biological Survey and the Department of Ecology and Evolutionary Biology at the University of Kansas. From processing our molecular samples to bioinformatics and Linux, phylogenetic tree construction, and sequence-related statistics, I learned a great deal from those around me. After four years of work, countless edits, and seven submissions, our manuscript was ready (Our article here!).

Figure 1. Josh and I working on a project collecting plant seeds in the field.
In the end, we found that pathogens respond to climate gradients in undisturbed sites, and were less responsive in human impacted, disturbed sites. In undisturbed grasslands, precipitation and temperature gradients were important predictors of both pathogen community richness and composition. Oomycete richness increased with precipitation, while fungal pathogen richness depended on an interaction of precipitation and temperature, with precipitation increasing richness most with higher temperatures (Fig. 2). Disturbance altered plant pathogen composition and precipitation and temperature had a reduced effect on pathogen richness and composition in disturbed grasslands.
Because pathogens can mediate plant community diversity and structure, the sensitivity of pathogens to disturbance and climate suggests that degradation of the pathogen community may mediate loss, or limit restoration of, native plant diversity in disturbed grasslands, and may modify plant community response to climate change. Our results highlight the importance of considering indirect mediators of plant community responses to environmental and land use changes. In addition, the interaction between precipitation and temperature in predicting fungal pathogen diversity underscores the importance of studying shifts in precipitation and temperature simultaneously, since looking at only one will not tell the whole story.
This manuscript is the result of a large amount of troubleshooting, trying, failing and trying again, as well as community support and guidance. We are very excited to have this paper published in ISME and hope this work will spur further research about the role of pathogens, and plant-associated microbes more generally, in mediating plant community responses to climate and land use changes.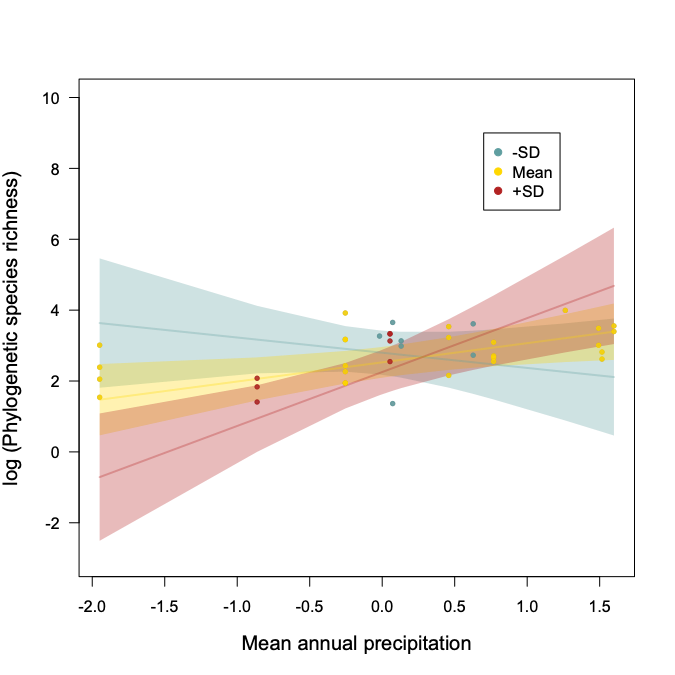
Figure 2. The interaction between precipitation and temperature in predicting fungal pathogen phylogenetic species richness (PSR). PSR increases with precipitation only under higher than average temperature.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in