Injecting molecules one at a time into cells.
Counting how many biomolecules are delivered into a cell?
Manipulating cells through the introduction of biomolecules into cells is routine in biology labs. Introducing biomolecules into living cells has allowed researchers to study many different cellular processes in normal physiology and in disease. These biomolecules can include DNA plasmids, small functional RNAs, CRISPR complexes, and proteins.
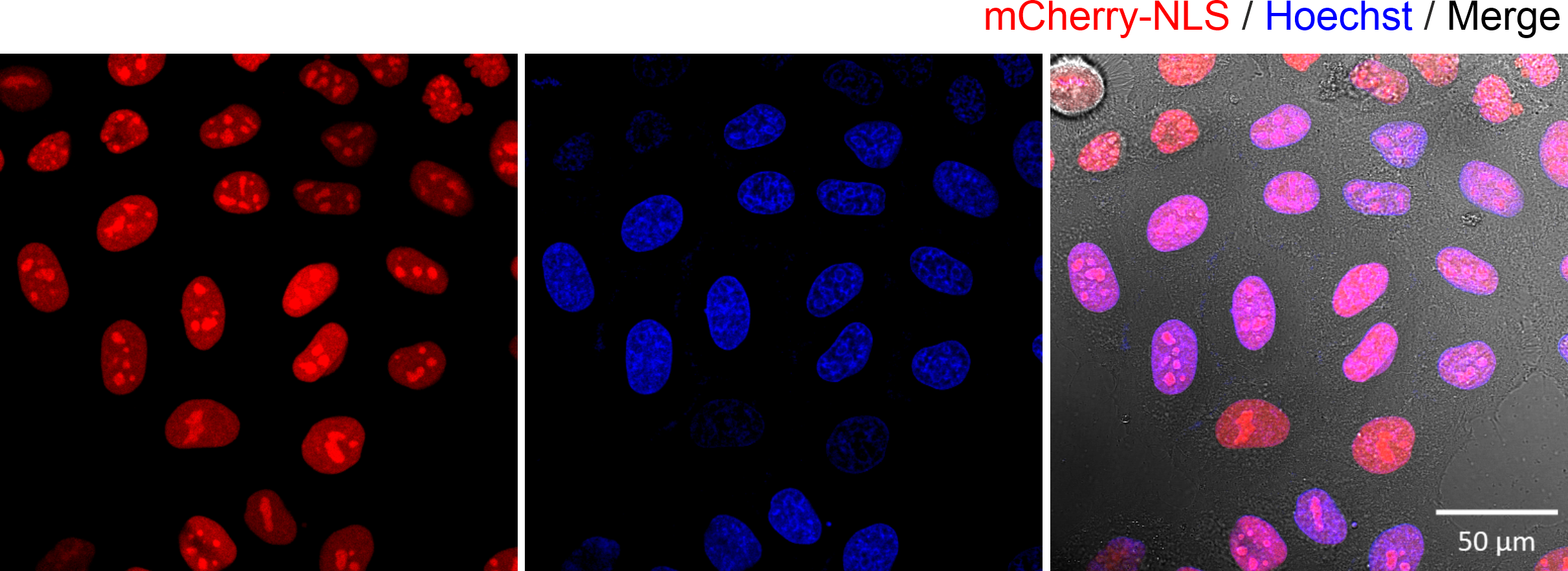
Figure 1. Fluorescence microscopy images of HeLa cells expressing the mCherry-NLS protein. The expressed proteins were found in the nucleus (co-localised with the Hoechst DNA dye). The transfection was carried out using lipofection transfection.
In the Figure 1 above, we employed lipofection, a method commonly used by cell biologists to transfect cells. In the experiment, a DNA plasmid, encoding the mCherry protein fluorescent fused with a sequence encoding a nuclear localization signal, was encapsulated inside lipid nanoparticles. Upon addition to the cells, the lipid nanoparticles then fused with the cell membrane, subsequently releasing plasmids into the cell cytoplasm. The results were clear: the cells expressed mCherry fluorescent proteins that were localized within the nucleus. While this method is effective, it has a significant limitation. We do not know how many plasmids were delivered into the cell. This means we cannot use this method to quantify how many biomolecules are required to elicit a cellular effect, in this case expression of protein in the cell’s nucleus. In order to quantify directly the number of biomolecules delivered into a cell, a different approach is needed. For this we developed the single-molecule nanoinjection platform.
Quantitative delivery of biomolecules through nanoinjection.
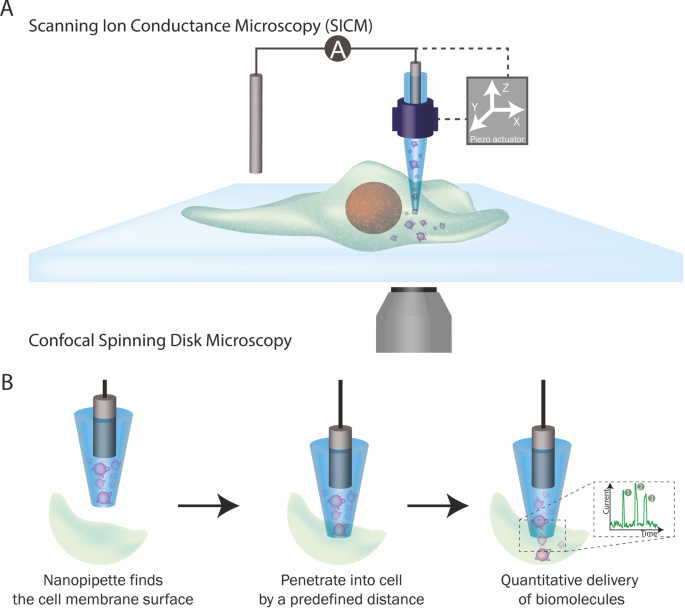
Two technologies are incorporated into this platform: SICM and nanopore single-molecule sensing. SICM is a type of scanning probe microscopy technique that uses a glass probe, called a nanopipette, to map the surface topography of substrates, such as the cell surface. The operation of SICM relies on ionic current as feedback: when the nanopipette approaches a surface, the current drops and this information can be used to generate the topographical profile of a cell.
Nanopipettes can also be used as a solid-state nanopores When the nanopipette pore opening is less than 100 nm in diameter, it can be used to perform single-molecule detection at single-molecule resolution. The voltage-driven translocation of biomolecules through the nanopore elicits a single current response, typically visualized as an ionic current peak. Each peak corresponds to the translocation of a single molecule across the nanopore meaning that by counting the number of peaks, you can quantify the number of biomolecules that have translocated through the nanopore.
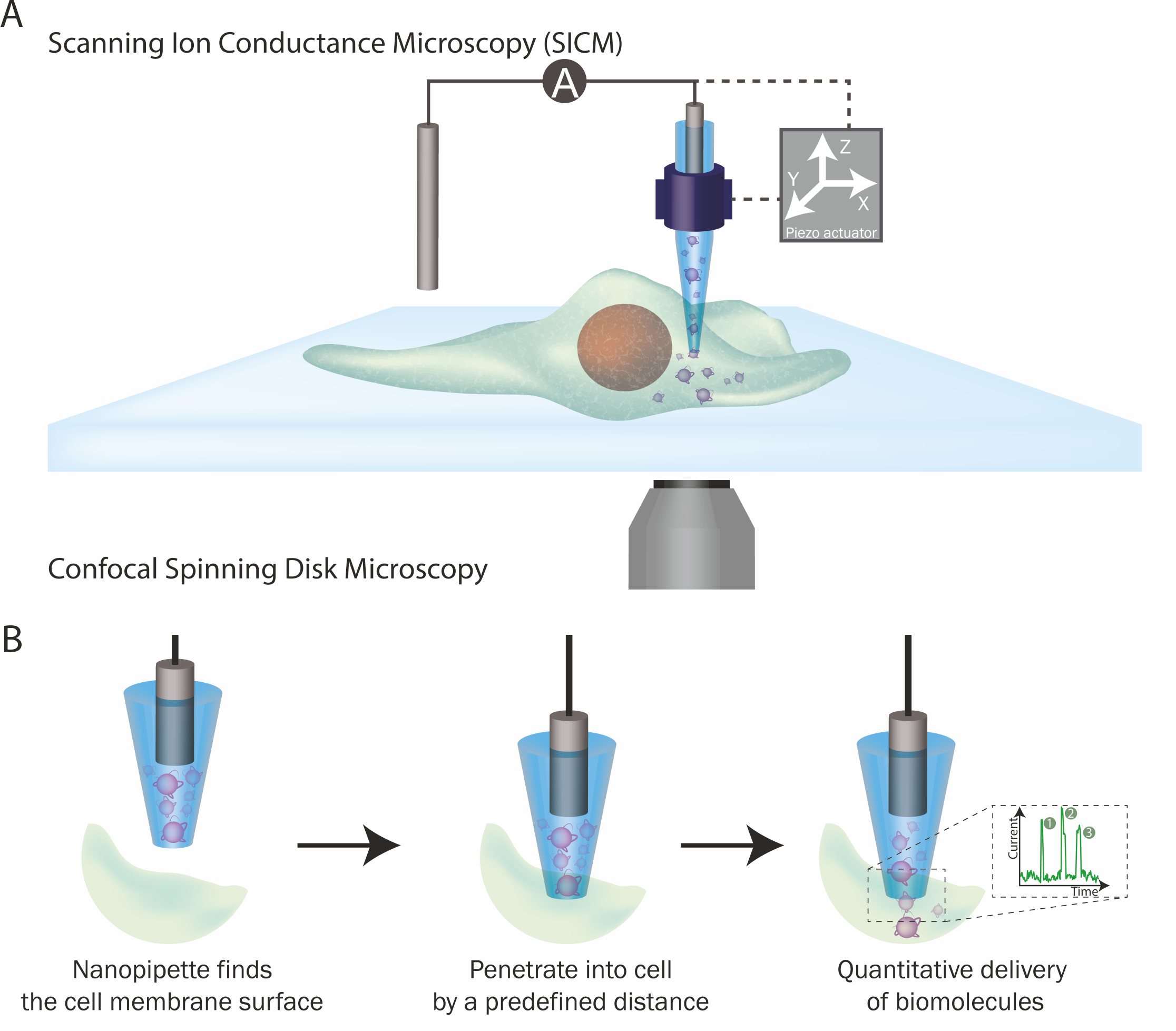
Figure 2. The single-molecule nanoinjection platform. (A) SICM integration into the platform. The position of the nanopipette is controlled by the SICM. (B) The quantitative nanoinjection procedure. The nanopipette approaches the surface of the cell membrane through the spatial control of the SICM, then the nanopipette is moved downward by a predefined distance to penetrate the cell, finally, the delivery of biomolecules will be triggered by electrophoretic forces via the application of a suitable voltage. During delivery, the current is monitored in real time and the translocation of a single analyte disrupts the current baseline and appearing as a peak, counting the number of peaks thus reveals the number of molecules delivered to the cell.
The integration of these two technologies led to the development of the single-molecule nanoinjection platform. A nanopipette loaded with biomolecules, is used as the SICM probe to approach a cell's surface and is then inserted into the cell at a defined depth. Once inserted, the voltage-driven translocation of each individual biomolecule from the nanopipette through the nanopore into the cell, generating a single ionic current peak. By quantifying the number of ionic current peaks, we can calculate the number of biomolecules delivered into the cell. Thus, the phenotypic effects of the biomolecules can then be correlated with the number of biomolecules delivered.
In our article, we employed single-molecule nanoinjection to transfect cells one at a time and then quantified the number of DNA plasmids delivered. In the figure below we loaded the nanopipettes with plasmids encoding a bright GFP variant, through the SICM, we inserted the nanopipettes tip into the nucleus of the cell, then a voltage was applied to cause the DNA plasmids to translocate from the nanopipette into the nucleus. Through counting the number of ionic current peaks, we quantified the number of DNA plasmids translocated into the nucleus and showed that fewer than 132 and 41 DNA plasmids were sufficient to transfect HeLa cell and primary neuron respectively such that GFP was expressed.
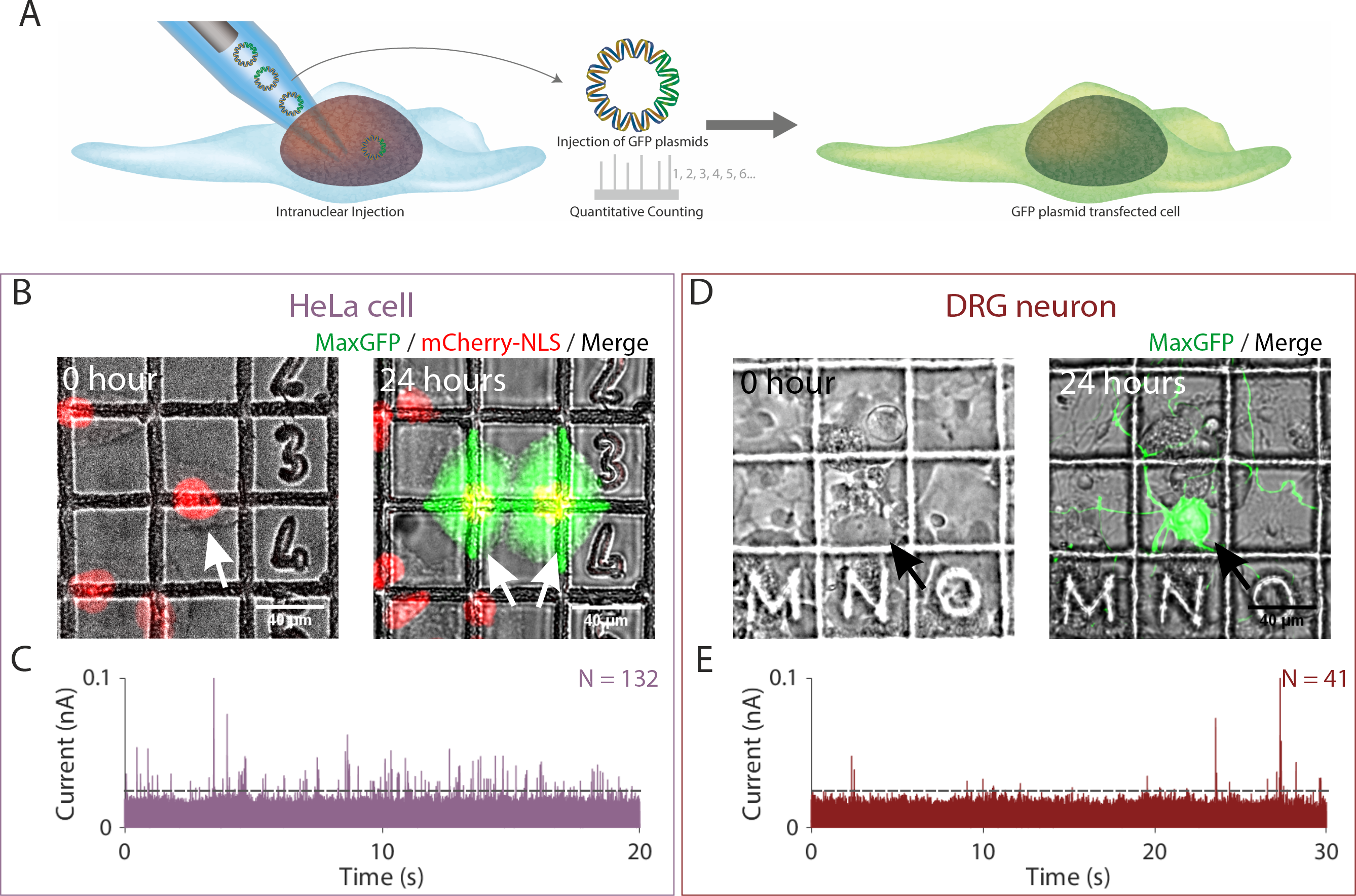
Figure 3. Quantitative nanoinjection of DNA plasmids into living cells. (A) Schematic of the nanoinjection of GFP plasmids (pMaxGFP) into the nucleus and the transfection of the cell. (B) The transfection of a HeLa cell expressing the nuclear localised mCherry-NLS (HeLa RNuc) with pMaxGFP plasmid through quantitative nanoinjection. HeLa RNuc cells were cultured on a grided dish to enable identification of the cell after nanoinjection. pMaxGFP plasmids were quantitatively nanoinjected into the nucleus of the cell (arrow). 24 hours later, the two daughter cells were imaged to confirm the expression of GFP from the injected pMaxGFP plasmids. (C) A snapshot of the current trace (20 seconds) recorded during the nanoinjection. Based on peak counting, a total of 132 pMaxGFP plasmids were nanoinjected into the HeLa RNuc cell. (D) The transfection of a DRG primary neuron with pMaxGFP plasmid through quantitative nanoinjection. 24 hours later, the neuron was imaged to confirm expression of GFP. (E) The current trace (20 seconds) recorded during the nanoinjection step. A total of 41 pMaxGFP plasmids were delivered into the DRG neuron. The dotted line in (C) and (D) indicated the threshold for events search.
The cell increases the sensitivity of the nanopore.
Unexpectedly, we observed that the intracellular environment improved the sensitivity of the nanopore, increasing the signal to noise ratio (SNR) during the translocation of biomolecules into cells, enhancing the detection of biomolecules delivered into cells. This enhancement of the SNR was reproduced if DNA was translocated into an electrolyte bath containing 30% (w/v) bovine serum albumin (BSA), which mimics the highly crowded intracellular environment.
To understand the mechanism of this enhancement, we utilized molecular dynamics simulations to model the translocation of DNA with and without crowder 30% (w/v) BSA. Modelling revealed that the translocation of DNA into BSA was slower, and the DNA molecule remained closer to the nanopore when compared to DNA translocated in the absence of BSA. These two factors contributed to the increased sensitivity of the nanopore. More on this can be found in our article.
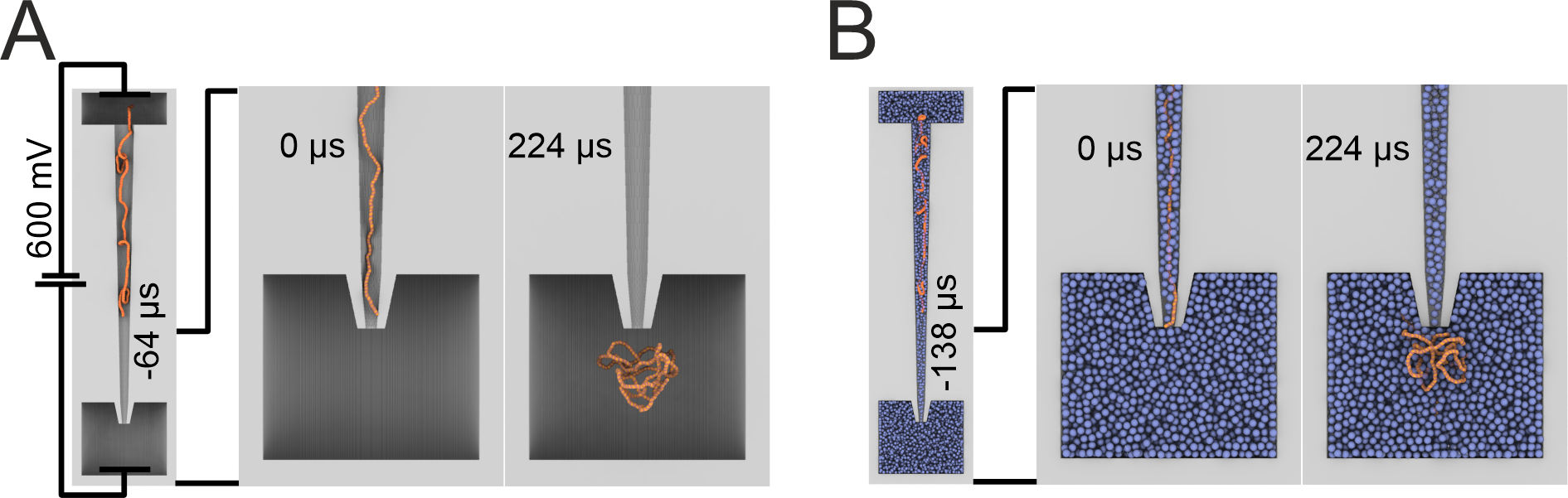
Figure 4. Coarse-grained simulation systems consisting of a 2.7 kbp dsDNA molecule (orange) driven out of a nanopipette (gray) by an applied electric potential into an electrolyte solution without (A) and with BSA (B) (blue). Each case (with or without BSA) consisted of an ensemble of 24 independent simulations.
Potential applications for nanoinjection for cell biology.
Whilst, thus far we have focused our discussion here on the delivery of DNA plasmids to transfect cells, in our article we also show that nanoinjection can deliver proteins into cells. We quantified the delivery of an enzyme, beta-galactosidase, and alpha-synuclein amyloid fibrils into different cell types thus demonstrate that this platform can be applied to a wide range of biomolecules. This we believe opens up a wide variety of applications for our nanoinjection platform. For example, misfolded proteins can aggregate into amyloid fibrils in neurodegenerative diseases, such as Parkinson’s. The physical properties of these fibrils, such as aggregation kinetics, seeding kinetics, and structure, can be extensively studied using different biophysical and biochemical techniques. However, the behaviour of amyloid fibrils inside a cell is more challenging to study. Our platform not only enables the intracellular delivery of biophysically characterised fibrils into neurons, but it will also enable the number of fibrils required for phenotypic effects such as seeding aggregation of cellular proteins into aggregates or to induce cell stress and death to be determined. Thus, with nanoinjection we can study cellular effects of biomolecules with single molecule precision.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in