Protecting Antibody IP Using Paratope Mapping & CDR-Scanning
Published in Bioengineering & Biotechnology and Business & Management
When monoclonal antibodies were first isolated in the 1970s, scientists lacked the tools to fully describe their structure. So, antibody patents mostly focused on function, claiming the ability of an antibody to bind a particular target protein or region of the protein. This approach provided developers with broad patent protection under previous patent guidelines. However, with over 200 therapeutic antibodies now approved and a global market worth hundreds of billions of dollars, such expansive patents have faced increasing scrutiny. A key turning point came with the 2023 U.S. Supreme Court case Amgen v. Sanofi, which invalidated broad functional claims in the U.S. With broad functional claims no longer an option, antibody developers have been left wondering how to protect their valuable Intellectual Property (IP), with some expressing concern that perhaps there is no longer a way to broadly protect antibodies.
At Integral Molecular, our scientists have spent decades expressing, mapping, and engineering antibodies and their targets. With a change in patent guidelines, we realized that we could apply our high-throughput protein engineering platform to fully characterize antibodies for IP applications, thereby closing this patent strategy gap.

The Amgen v. Sanofi1 Supreme Court case centered around a functionally described genus of antibodies targeting a region of PCSK9, a drug target for lowering cholesterol. Although the patent listed 26 specific antibodies, it was deemed invalid because it did not adequately teach how to make the full claimed genus. Following the decision, guidance2 from the United States Patent and Trademark Office (USPTO) clarified that patent applications must meet the “enablement” requirement—meaning they must provide sufficient detail for someone skilled in the art to make and use the invention without undue experimentation.
Based on the new guidance, it is clear that antibody patents must both enable the invention (describe it in sufficient detail) and provide written description (demonstrate possession of the claimed invention). However, none of the USPTO guidelines or court cases outline specific technical strategies to meet these requirements.
The Solution: Claiming an Antibody Structural Genus
Scientists at Integral Molecular collaborated with several patent attorneys, including coauthor Ulrich Storz, to develop an innovative solution, CDR-scanning, to meet the USPTO’s enablement and written description requirements while offering robust protection for antibody IP. We describe this strategy in Banik et al., 2025, Nature Biotechnology.
Drawing on recent legal developments, case law, and available technology, we developed a strategy to claim a structural genus of antibodies that mirrors the strategy used in small-molecule drug patents. Here, a core scaffold structure is defined along with permissible substitutions that preserve the molecule’s function. Applying this approach to antibodies, the antibody paratope (the amino acids that contact the target protein) defines the core common structure of the genus of antibodies. The amino acid substitutions in the antibody’s CDR region that permit target binding constitute the permissible substitutions. The parent antibody and permissible variations then represent a genus that can be claimed.
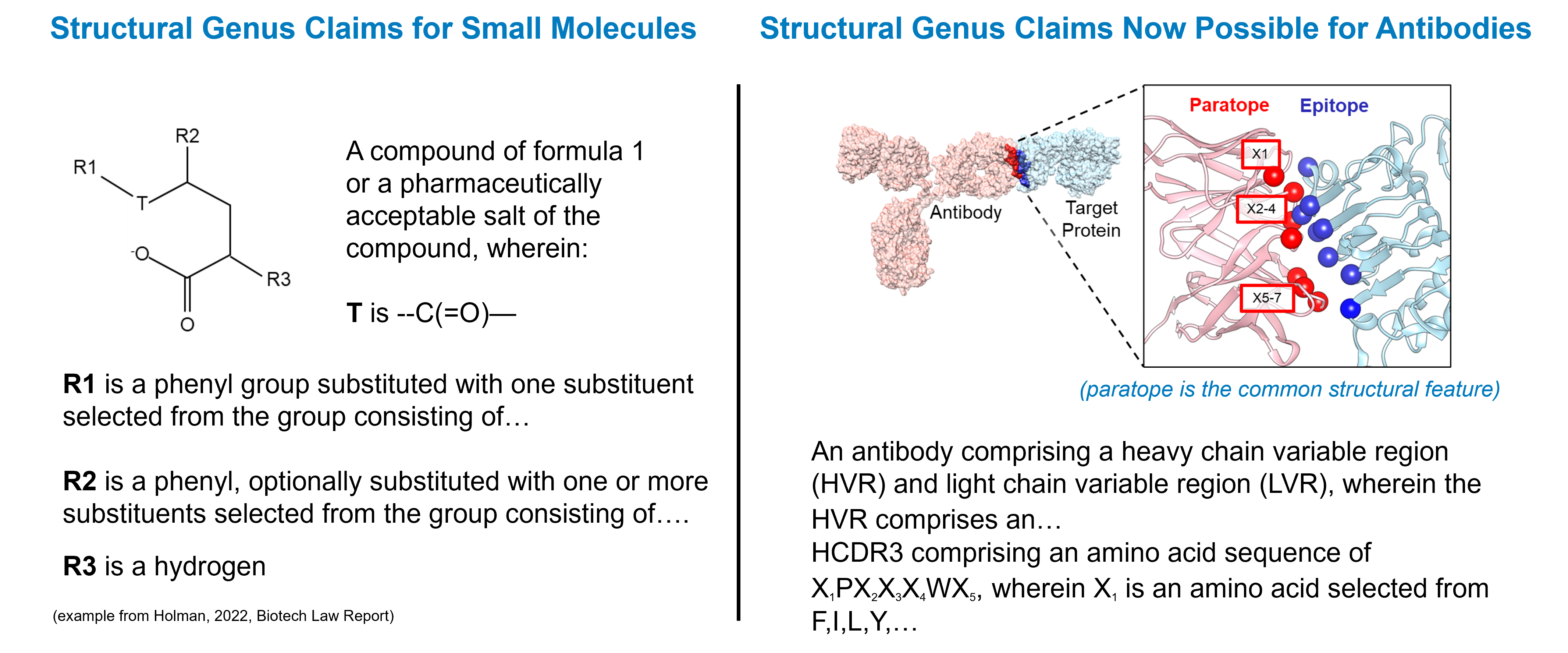
What Does CDR-Scanning Entail?
The CDR-scanning strategy involves comprehensive mutational scanning of the variable CDR residues of the antibody (typically ~50-60 residues), where each position is mutated to all 19 other possible amino acids. These variants are all tested for the ability to bind the therapeutic target. Using modern high-throughput experimental methods, it is now technically, economically, and logistically feasible to make and test this number of variants.
How CDR-Scanning Fulfills Enablement and Written Description Requirements
CDR-scanning enables inventors to:
- Construct claims for a parent antibody and genus of related antibodies that share a common paratope structure
- Demonstrate both enablement and written description of the genus of antibodies in their possession
- Write broad patent claims to protect their antibody
For proper enablement, the patent application must describe the invention in sufficient detail such that someone with ordinary skill in the art can “make or use” the claimed invention. Furthermore, it should describe some “general quality running through the genus that gives rise to the claimed function.” CDR-scanning provides this information, highlighting the critical paratope residues (the common structural feature) and permissible substitutions.
The written description must include enough details and examples to demonstrate possession of the full scope of the invention at the time of filing. Because CDR-scanning involves experimentally cloning and testing each variant, it clearly fulfills the written description requirement.
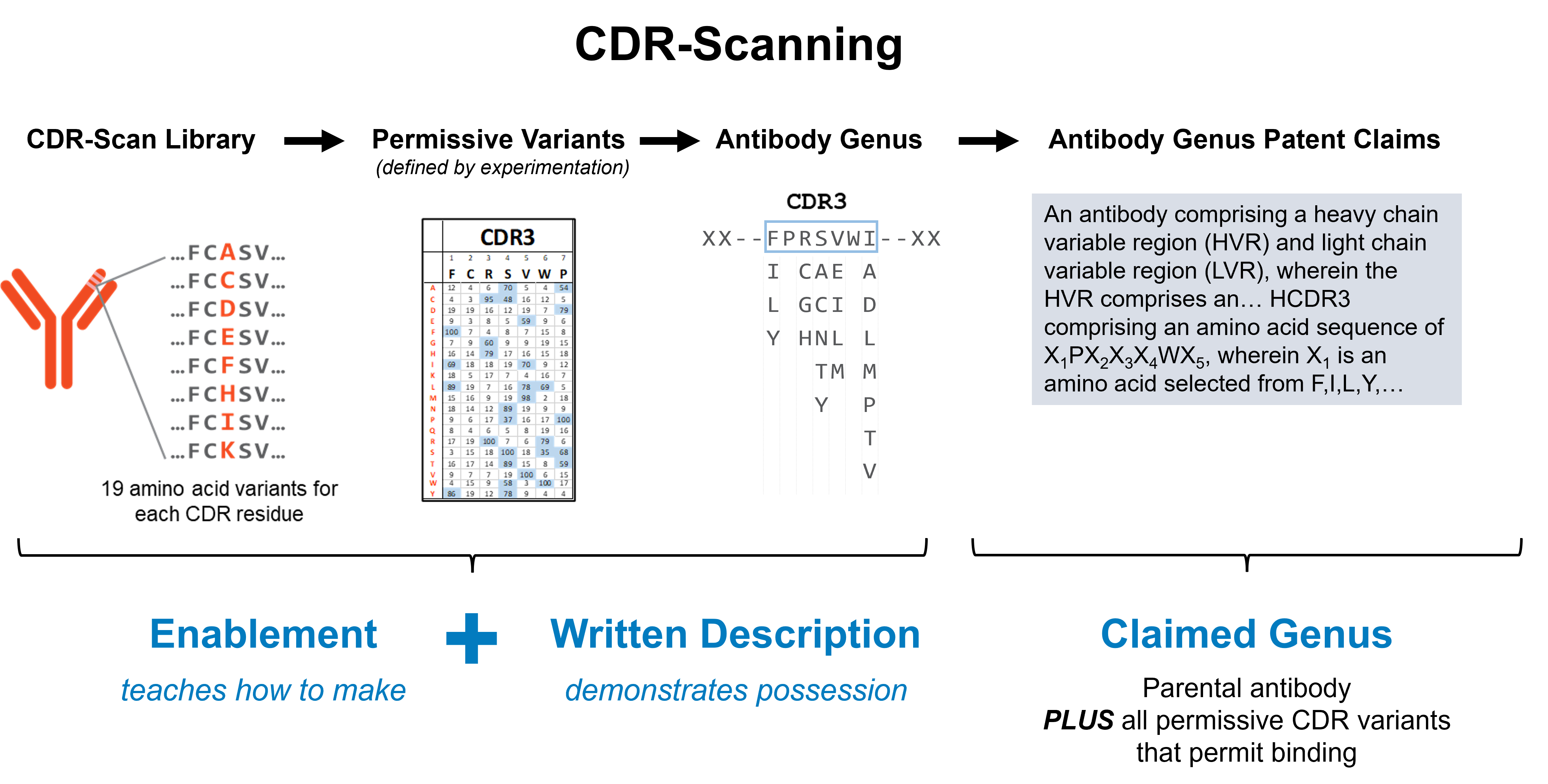
CDR-Scanning Enables Antibody Engineering
CDR-scanning also provides valuable data for engineering antibodies that serves as the basis for additional novel IP and patents in addition to the original claims. Improvements revealed by CDR-scanning can include:
- Increased binding to the target of interest
- Increased antibody production
- Better developability
As the rules around antibody patents continue to evolve, a strategy based on paratope mapping and CDR-scanning could be the key to securing strong IP. By combining modern antibody technologies with a clear understanding of the current legal requirements, antibody developers can better protect their IP while also expanding scientific innovation around their antibody sequence.
References
1. Amgen, Inc. v. Sanofi, 598 US 594 (2023).
2. US Patent and Trademark Office. Guidelines for Assessing Enablement in Utility Applications and Patents in View of the Supreme Court Decision in Amgen Inc., et al. v Sanofi et al. (10 January 2024).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in