Single cell histone modifications can be readily quantified in single cell proteomic datasets
Published in Protocols & Methods and Genetics & Genomics
Single cell proteomics (SCP) by liquid chromatography mass spectrometry (LCMS) is rapidly increasing in both throughput and proteomic coverage per cell. Like all LCMS methods, detection is biased toward the highest concentration proteins. In 2022 we (Orsburn et al., Nature Comms) demonstrated the detection and relative quantification of protein post-translational modifications (PTMs) on very high abundance proteins in single cells. We didn't overlook the fact that most of these were on nuclear histone proteins.
To follow up on this, that summer I presented a lightning talk and poster at the Johns Hopkins Epigenome Cluster annual meeting in an attempt to determine whether this would be an interesting avenue of research for the epigenetics heavy hitters in attendance (I'm in the orange backpack with the cool organic chemistry tie). My single slide was basically showing all the histone PTMs we could detect with SCP at that time and an open invitation to discuss collaborations at my poster conveniently near the back corner fire escape. A couple students dropped by looking for jobs, but not one researcher in attendance took me up on a conversation. I generally pursue ideas until someone convinces me that the idea is not worth pursuing, and since no one talked me out of it, I did a solo weekend and evening project on it.
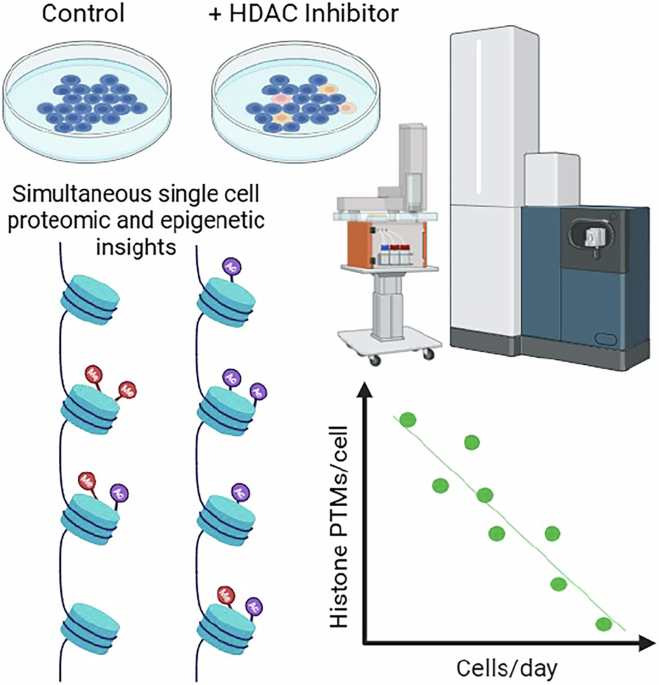
I took some cancer cells I knew how to grow effectively and I treated them with a histone deacetylase inhibitor that people were recently publishing on (mocetinostat), randomized control and treated cells and performed SCP at different throughputs. As throughput goes up, SCP (and all proteomics) coverage generally goes down. Since I was focusing on proteins with millions of copies per cell, I figured I could get away with far higher throughput than my previous studies. And, for the most part, I could. In our standard SCP workflow described above we analyzed around 210 cells per day. In the the mocetinostat study I tried methods with throughputs as high as 3500 cells per day. While histone proteins could be detected at more than 700 cells per day, the quantitative accuracy was not acceptable, and there weren't more than a handful of confident PTMs identified. However, 700 cells per day was a tremendous increase in our throughput and could identify multiple histone PTMs in each human cell with only minor compromises in quantitative accuracy. Surprisingly, I could still quantify hundreds of unrelated proteins in every cell anyway, so either histone PTMs or whole proteins could be considered a free consequence of the other motive.
I dropped the files on a public repository and a quick preprint where I focused on the throughput, because it didn't seem anyone was all that interested in the biology.
In January of 2024 I was invited to give a workshop at the amazing EuBIC Winter School and discovered that researchers at Albert Einstein in collaboration with the Broad were also diving into single cell histone PTMs with targeted methods empowered by picoliter scale robotics. I'll be honest, I did not believe this was even remotely possible, even though I had the same robot and instrument. A fantastic talk by Lauren Corveleyn proved me wrong. If Simone Sidoli and Steve Carr are interested in a topic, in my mind that absolutely means there must be a practical application for it and I should probably dig into my data a bit more. The method Lauren described in Winterberg turned into an amazing work (Cutler, Corvelyn and Ctortecka et al.,) that published in Nature Communications just a few weeks ahead of mine this week in Communications Biology.
Is there a future for single cell PTM analysis? Does anyone really want to know what dozens of histone PTMs are doing in single cells? That's up to the biologists and clinicians. I'm a mass spectrometrist who works best as a cog in a collaborative wheel. What I do know, is that if there is an application, two complementary technologies now exist and both the Sidoli lab and my lab (and the Pitt Health Sciences Mass Spectrometry core facility) are very open to discuss collaborations in this space.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in