The ribosomal state plasticity is essential to forming the tumor-permissive microenvironment
Published in Cancer
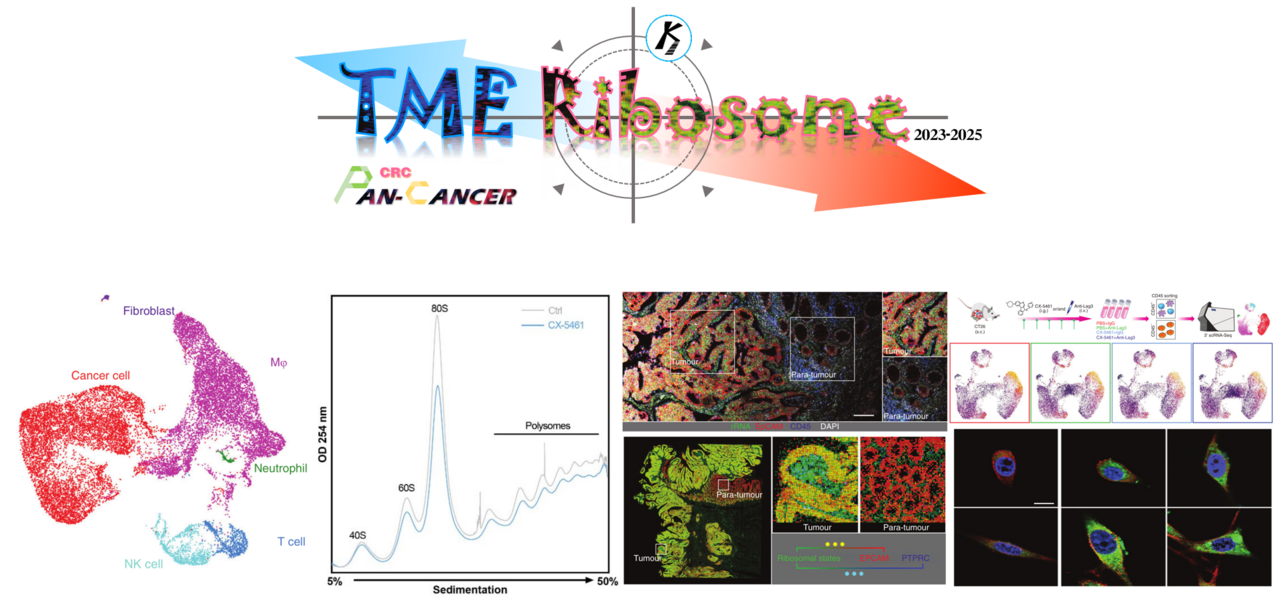
Hyperactive ribosome biogenesis in cancer cells is a hallmark of tumor initiation and progression. Hence, current ribosome-related studies, including our previous studies, were concentrated on cancer cells. The rRNA metabolic process is critical for protein synthesis and increased protein synthesis is required for tumorigenesis. We have comprehensively characterized rRNA metabolism-related genes in human cancer and further investigated proliferation-prompting functions and mechanisms of an rRNA metabolism-related gene EXOSC8 [1, 2] (EXOSC8 is an rRNA metabolism-related oncogene in CRC).
Actually, ribosomes are among the oldest molecular machines in extant life and translate information from mRNAs into functional proteins within cells, of course including non-cancer cells. Recently, a series of reports showed that partly non-cancer cells also contributed to protumorigenic tumor microenvironments (TME). We therefore asked these questions in our research: (1) Whether these “bad” non-cancer cells also need relatively hyperactive ribosome biogenesis than other non-cancer cells? (2) If (1) is true, what is the biological consequence of targeting ribosome biogenesis in “bad” non-cancer cells and is approach able to drive anti-tumor immunity?
However, traditional bulk transcriptome examination mixes gene expressions of overall cells in samples, resulting in limited and unclear information on expressions, proportions and interactions among cells. Fortunately, single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) technologies have improved our understanding of cancers by characterizing transcriptomes at a cellular resolution, identifying cell types and their expression profiles. A previous study used ACTIONet algorithm to identify epithelial-mesenchymal plasticity (EMP) programs and defined a conserved signature associated with EMP in scRNA-seq data [3]. In this study, we used the non-negative matrix factorization (NMF) approach to learn coordinated expression programs consistent with intertumoral ribosome biogenesis signaling at the single-cell resolution, developing a general signature to characterize the ribosome biogenesis (ribosomal state) of individual cells within the TME. The NMF approach was used in our previous studies in TME of solid tumors, such as colorectal cancer (CRC) [4], prostate cancer [5] and glioma [6] (Figure. 1).
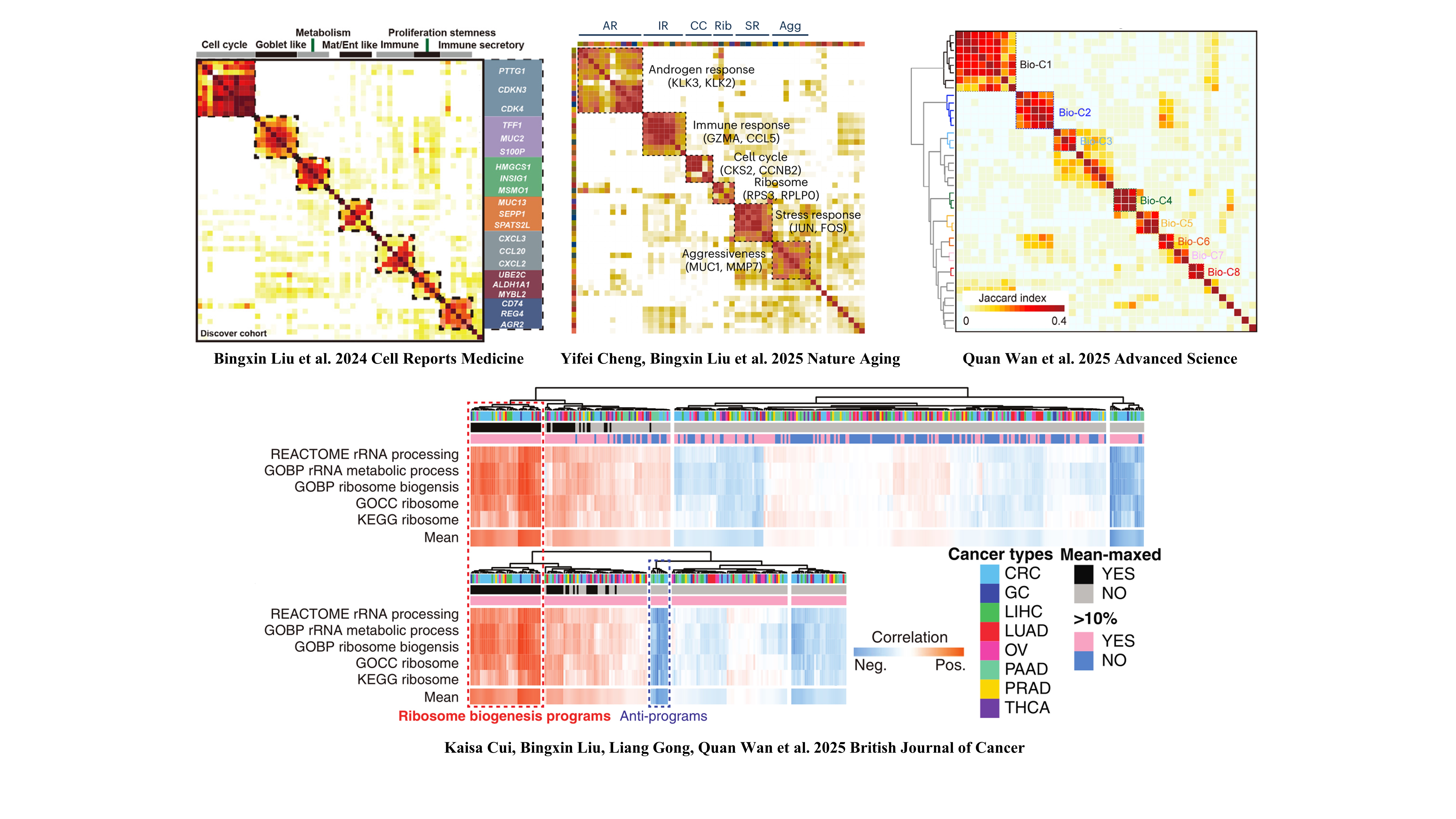
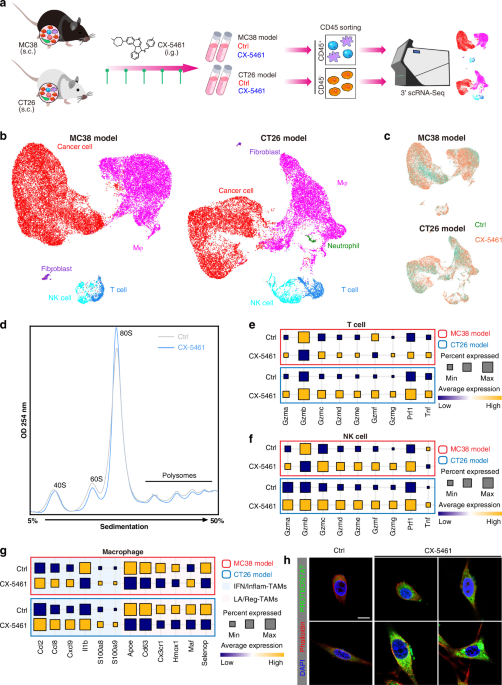
Figure 1. The NMF approach were used in our previous studies in TME of solid tumors
By using our 73-metagene ribosomal state signature and large-scale human Pan-cancer scRNA-seq data, we uncovered unique immune cell subpopulations with relatively active ribosomal state, including stress response CD8+ T cell (CD8+ Tstr), regulatory T cells (Treg), immunosuppressive-like and lipid-associated (LA)/ immune regulatory (Reg)- tumor-associated macrophages (MRC1+ TAMs). These cell subpopulations were also associated with adverse clinical outcomes. We performed ribosome-targeting therapy utilizing CX-5461, an effective and acknowledged selective inhibitor of ribosome biogenesis, in immunocompetent in vivo models and submitted for scRNA-seq. In line with abovementioned findings in Pan-cancer scRNA-seq data, ribosome inhibition meaningfully regressed these cell subpopulations in vivo (Figure. 2A). Furthermore, ribosome inhibition elevated lymphoid cell cytotoxic granule secretion and macrophage pro-inflammation reprogramming (Figure. 2B), but induced immune checkpoint (IC) expression (such as Lag3). Targeting ribosomes significantly sensitizes tumors to anti-Lag3 immunotherapy, eliciting potent tumor regression and deeper anti-tumor immune responses (Figure. 2C) [7].
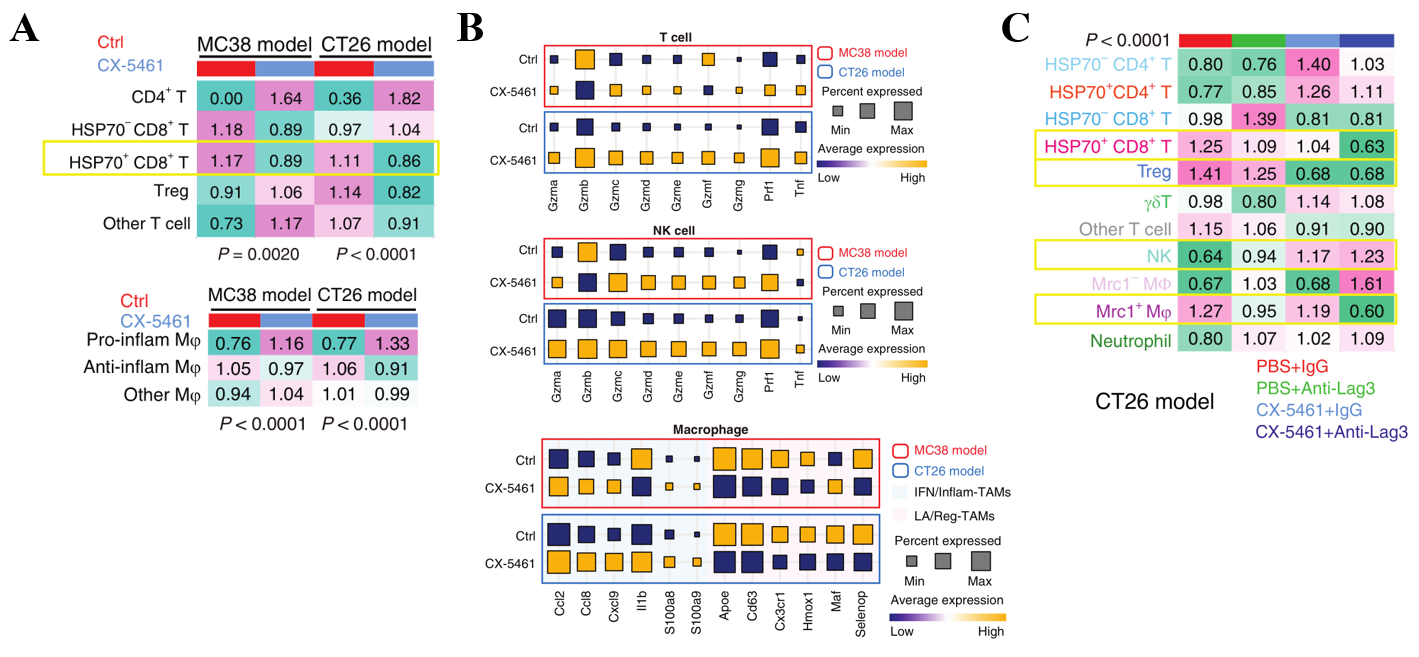
Figure 2. Unique immune cell subpopulations that are sensitive to ribosome biogenesis blockade.
We have discussed some interesting points in the published manuscript. In this blog, we would like to further discuss four points. (1) Ribosomal state metagenes contain several canonical ribosome-related genes. We noted that canonical ribosomal protein (RP) genes are not included in this signature [7]. Thus, our ribosomal state metagenes are not be disrupted by filtering (mitochondrial) ribosomal L/S-protein genes when single-cell data quality control is needed. For instance, although (mitochondrial) ribosomal genes and several immune genes were excluded from Visium HD (FFPE v1 technology) data, our ribosomal state signature can be used in these Visium HD data. Indeed, we also performed Visium HD in other ribosome projects, and this ribosomal state helped us to explore the correlation between related genes and ribosome biogenesis level with single-cell-scale resolution and spatial accuracy.
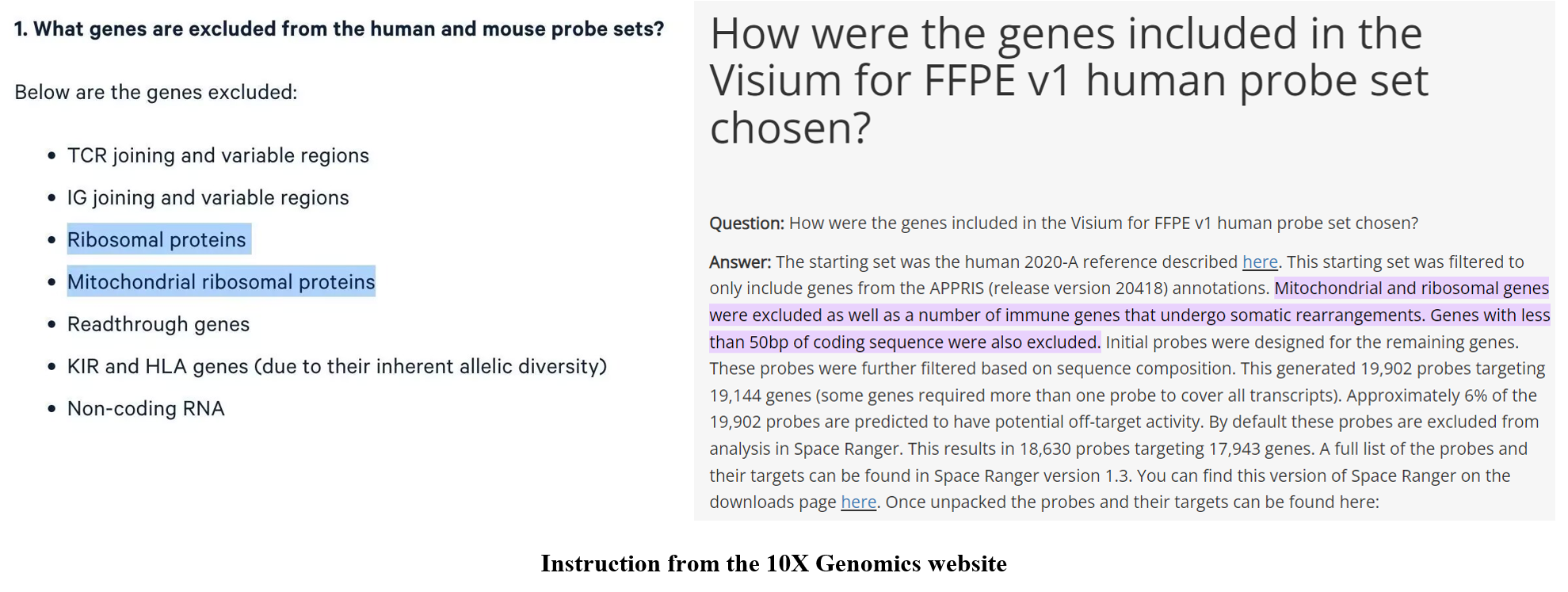
Figure 3. Our ribosomal state signature can be used in these Visium HD data
(2) Unique “hot” ribosomal state cell subpopulations may serve as a therapeutic vulnerability in protumorigenic and immunosuppressive TME. Global ribosome blockade promotes anti-tumor immune responses in vivo, but also induces expression of some ICs [7]. These features seem similar to immune overdrive TME phenotypes reported in our previous studies, which may exhibit better responses after ICB [8, 9] (Immune overdrive high-risk subpopulations in CRC and New insights into CTSL/ZBTB7B in gastric cancer prognosis and TME).
(3) After submitting this manuscript, we found newly published papers that confirmed our findings. Metge et al. uncovered that rRNA synthesis was increased in tumor-supportive macrophages, providing evidence that ribosome biogenesis is a targetable dependency to reprogram the tumor immune microenvironment [10]. Another study performed by Weller et al. showed that translation dysregulation in cancer as a source for targetable antigens [11], similar to our findings and the reviewer’s suggestion [7]. These studies further enhanced our confidence in this research.
(4) This study is the first time used pseudo immunofluorescence method that we developed [7]. This idea inspired by multi-color immunostaining in tissues. The color intensity of each cell (spot) represents the level of the corresponding gene/signature, while the multiple overlapping colors of each cell (spot) represents whether genes/signatures are co-expressed. Hence, this method is useful to analyzing ST data.
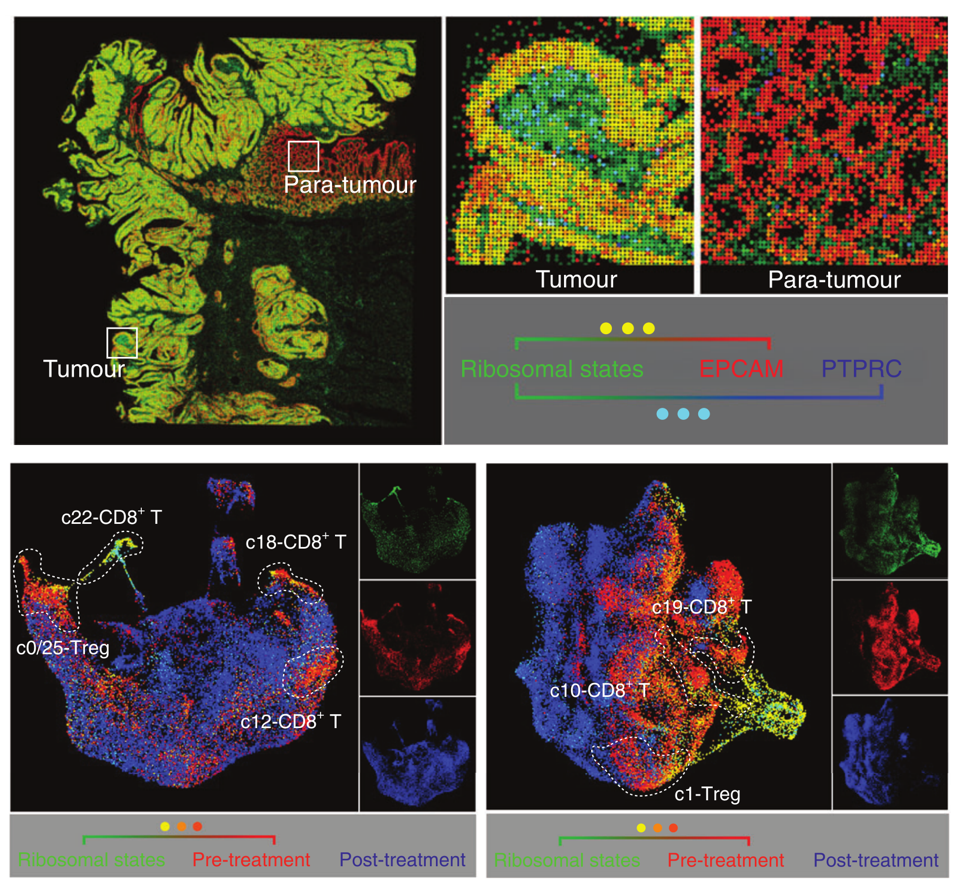
Figure 4. Pseudo immunofluorescence based on Visium HD and scRNA-seq in this study.
In conclusion, this study is the first to provide a single-cell atlas and potential mechanistic insight into ribosome-targeting therapy for reshaping the TME and enhancing immune checkpoint blockade (ICB) efficacy in pre-clinical models. This work further extends our previous studies on tumor ribosome biogenesis. Our work unveils that ribosome biogenesis blockade could reinstate immunosurveillance and provide novel strategies to enhance the ICB efficacy in patients with poor immunogenicity.

References
1 Cui K, Gong L, Zhang H, Chen Y, Liu B, Gong Z et al. EXOSC8 promotes colorectal cancer tumorigenesis via regulating ribosome biogenesis-related processes. Oncogene. 2022; 41: 5397-5410.
2 Cui K, Liu C, Li X, Zhang Q, Li Y. Comprehensive characterization of the rRNA metabolism-related genes in human cancer. Oncogene. 2020; 39: 786-800.
3 Cook DP, Vanderhyden BC. Transcriptional census of epithelial-mesenchymal plasticity in cancer. Sci Adv. 2022; 8: eabi7640.
4 Liu B, Li S, Cheng Y, Song P, Xu M, Li Z et al. Distinctive multicellular immunosuppressive hubs confer different intervention strategies for left- and right-sided colon cancers. Cell Rep Med. 2024; 5: 101589.
5 Cheng Y, Liu B, Xin J, Wu X, Li W, Shang J et al. Single-cell and spatial RNA sequencing identify divergent microenvironments and progression signatures in early- versus late-onset prostate cancer. Nat Aging. 2025; 5: 909-928.
6 Wan Q, Wu X, Zhou J, Wu W, Cao Y, Sun C et al. The Hypoxia-Associated High-Risk Cell Subpopulation Distinctly Enhances the Progression of Glioma. Adv Sci (Weinh). 2025; 12: e2416231.
7 Cui K, Liu B, Gong L, Wan Q, Tang H, Gong Z et al. Targeting ribosomes reprograms the tumour microenvironment and augments cancer immunotherapy. Br J Cancer. 2025.
8 Cui K, Yao S, Liu B, Sun S, Gong L, Li Q et al. A novel high-risk subpopulation identified by CTSL and ZBTB7B in gastric cancer. Br J Cancer. 2022; 127: 1450-1460.
9 Cui K, Yao S, Zhang H, Zhou M, Liu B, Cao Y et al. Identification of an immune overdrive high-risk subpopulation with aberrant expression of FOXP3 and CTLA4 in colorectal cancer. Oncogene. 2021; 40: 2130-2145.
10 Metge BJ, Williams L, Swain CA, Hinshaw DC, Elhamamsy AR, Chen D et al. Ribosomal RNA Biosynthesis Functionally Programs Tumor-Associated Macrophages to Support Breast Cancer Progression. Cancer Res. 2025; 85: 1459-1478.
11 Weller C, Bartok O, McGinnis CS, Palashati H, Chang TG, Malko D et al. Translation dysregulation in cancer as a source for targetable antigens. Cancer Cell. 2025; 43: 823-840 e818.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in