

The rRNA metabolic process is critical for protein synthesis and increased protein synthesis is required for tumorigenesis. rRNA metabolism-related genes are defined as genes associated with the chemical reactions and pathways involved in rRNA, such as ribosomal proteins (eg. RP5 and RPL11), DEAD/DEAH-box helicases (eg. DDX56), exosome components (eg. EXOSC8), small subunit processome components (UTP18), and WD repeat domain proteins (eg. WDR36) [1]. Moreover, to gain analytical breadth, and define commonalities, differences and emergent themes across human cancers, Pan-cancer analysis is beginning to be used in cancer research [2]. Thus, when I studied at Wuhan university (successive postgraduate and doctoral programs of study), I performed a Pan-cancer analysis of rRNA metabolism-related genes across 20 human cancers under the supervision of my Ph.D. supervisor (Prof. Li). A landscape of rRNA metabolism-related genes was established in human cancers [1]. We found that rRNA metabolism-related genes are upregulated in multiple cancers, particularly in digestive and respiratory system cancers. Most of the upregulated genes were driven by copy number variation (CNV) gain rather than mutation or DNA hypomethylation. We systematically identified CNV-driven rRNA metabolism-related genes with clinical relevance, including a critical candidate oncogene: EXOSC8, also named Rrp43p and OIP2. EXOSC8 is an essential protein of the exosome core, and the depletion of EXOSC8 results in a severe growth defect in yeast [3]. It was reported that EXOSC8 is necessary for the efficient maturation of 5.8S, 18S and 25S rRNA in S.cerevisiae [4]. Interestingly, EXOSC8 has not been reported in human cancer at that time. We therefore performed basic functional experiments in vitro and in vivo, including MTT assays and AOM/DSS-induced CRC mouse model. Our data indicate that EXOSC8 could serve as a valuable new biomarker and prognostic factor for CRC. This Pan-cancer study demonstrates that our analysis could be an effective resource to functionally identify rRNA metabolism-related genes with clinical relevance in human cancers. However, further functions and mechanisms of EXOSC8 in CRC were not clear, in particular its roles in ribosome biogenesis-related processes.
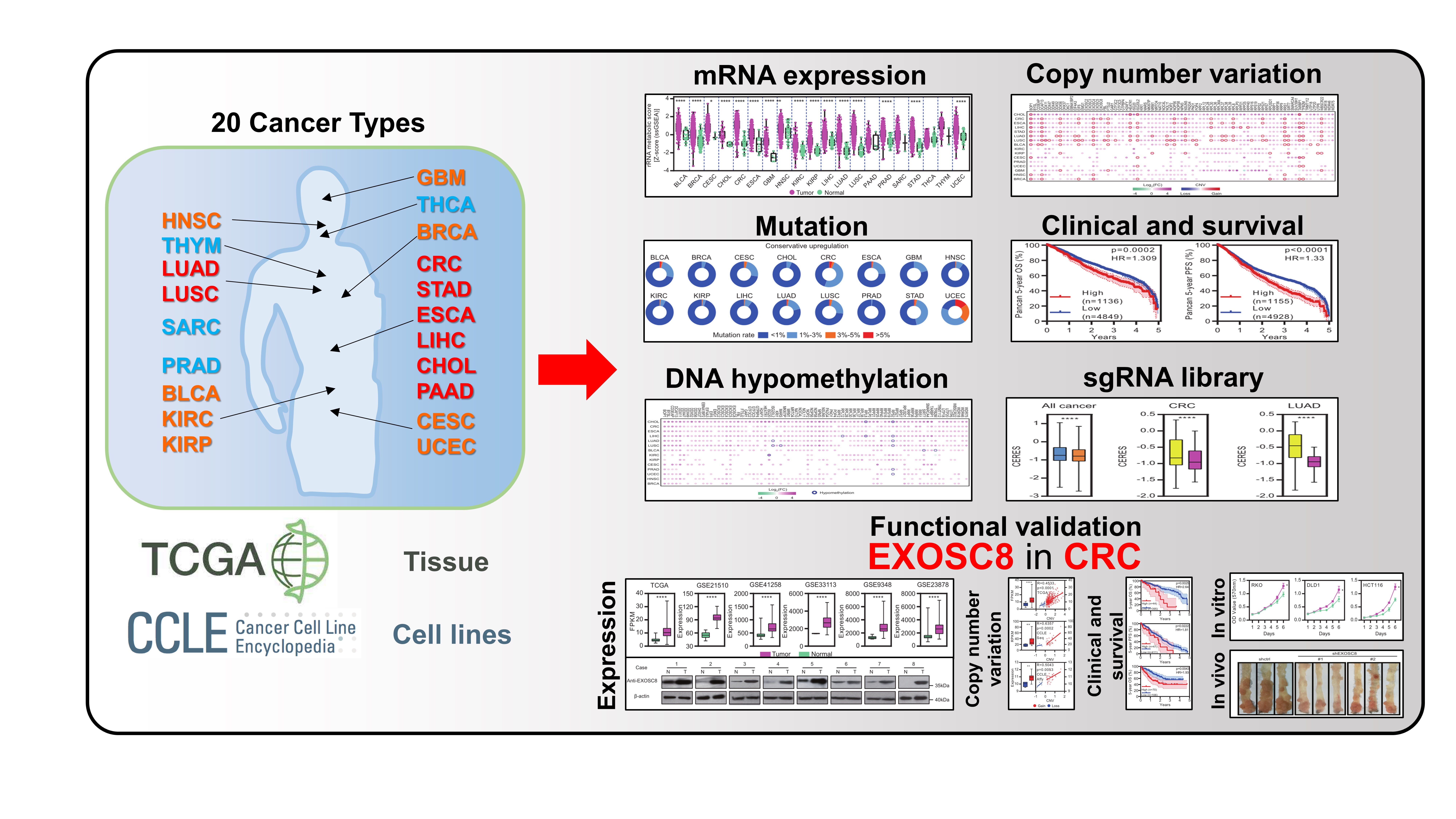
After I graduated from college, I joined in Prof. Huang’s team (Affiliated Hospital of Jiangnan University/Jiangnan University) in November 2019. He gives strong backing to my scientific research. Upon his support, we have published a series of studies about tumor microenvironment (TME), such as Immune overdrive high-risk subpopulations in CRC and New insights into CTSL/ZBTB7B in gastric cancer prognosis and TME [5, 6]. Apart from TME, he also supports my work to further investigated proliferation-prompting functions and mechanisms of EXOSC8 in CRC.

The common research logic of cancer-related genes is expressions→clinical survival→functions in vitro and in vivo→mechanisms (downstream and upstream). Recently, the availability of single-cell transcriptomic profiles across a broad range of human cancers provides an unprecedented opportunity to explore TME in greater depth [6, 7]. Thus, we firstly explored and verified the enhanced correlation between EXOSC8 and ribosome biogenesis-related factors/processes in CRC at the tissue, cell lines and single-cell levels. We originative used functional genesets (ribosome proteins, RPs; translation initiation factors, EIFs) to cluster epithelial cells (potential malignant cells) from CRC single-cell dataset, and uncovered that EXOSC8 was significantly co-expressed with RPs EIFs, proliferation marker genes and nucleolar protein genes. Then our wet-lab experiments confirmed that EXOSC8 expression is required for ribosome biogenesis-related processes in CRC. Increased ribosome biogenesis and protein synthesis play essential roles in sustaining tumor cell proliferation [8]. Our subsequent Pan-cancer analysis and wet-lab experiments reveal that EXOSC8 is overexpressed and required for CRC tumorigenesis. Besides, overexpression of EXOSC8 was positively correlated with CNV gain at the tissue, cell lines and single-cell levels.
The research logic and method of transcription factor and enzyme genes, etc. are clear and mature. However, EXOSC8, as an RNA exosome gene, how to find its mechanisms in CRC? Fortunately, first, our systemically and comprehensive biological analysis pointed out that EXOSC8 was meaningfully correlated with ribosome biogenesis-related processes in CRC; second, after our previously Pan-cancer study was published, a new study in 2020 suggested that EXOSC8 or EXOSC9 knockdown leads to a significant increase in p53 protein levels [9]; third, we noted that some rRNA metabolism-related genes, such as PNO1 and RRS1, could regulate RPs-Mdm2-p53 signaling in CRC and hepatocellular carcinoma, respectively [10, 11]. These findings motivated our next analyses and wet-lab experiments design. Our bioinformatic analyses and wet-lab experiments demonstrated that EXOSC8 knockdown increased p53 levels in CRC, and the oncogenic proliferation phenotypes of EXOSC8 depended on p53 in vitro and in vivo. Impressively, a series of protein-related assays suggested that when EXOSC8 is silenced, RPL5/11 would be released into the nucleoplasm and bind Mdm2, resulting in a decrease in the Mdm2-mediated ubiquitination and degradation of p53.
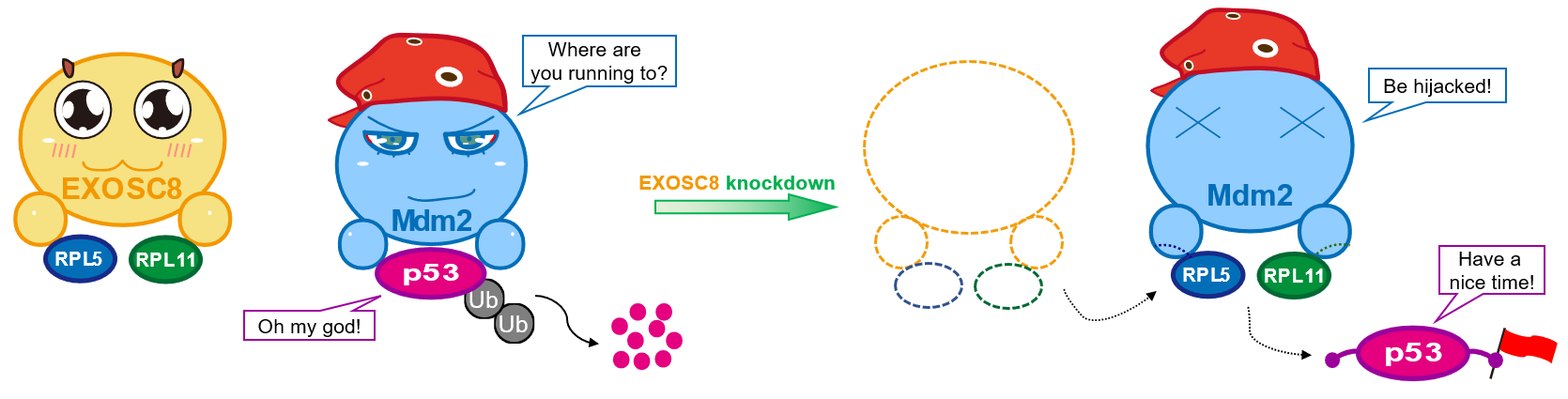
Thus, our combined computational and experimental analyses indicate that EXOSC8 alters the balance of RP5/11-Mdm2-p53 mediate ribosomal stress. EXOSC8 anchors RPL5/11 in the nucleus, particularly the nucleolus. EXOSC8-anchored RPL5/11 will be released into nucleoplasm from the nucleolus in CRC cells when EXOSC8 knockdown. This process impairs nucleolar and triggers ribosomal stress (nucleolar stress). On the one hand, the nucleus-released RPL5/11 interact directly with Mdm2 and blocks its E3 ubiquitin ligase function thereby releasing and accumulating p53. Upon activation, p53 transactivates several downstream targets, leading to cell cycle arrest (G1 arrest), etc. and controls CRC cells’ destinies. On the other side, impaired nucleolar contributes to perturbations in ribosome biogenesis, such as impaired rRNA processing and ribosome assembly, which decreases new protein synthesis and blocks the supply of materials for CRC cell proliferation. Importantly, according to Dr. Gong and Prof. Huang’s previous studies about therapy [12, 13], we performed a preliminary investigation and found that EXOSC8 might serve as a potential therapeutic target in CRC.
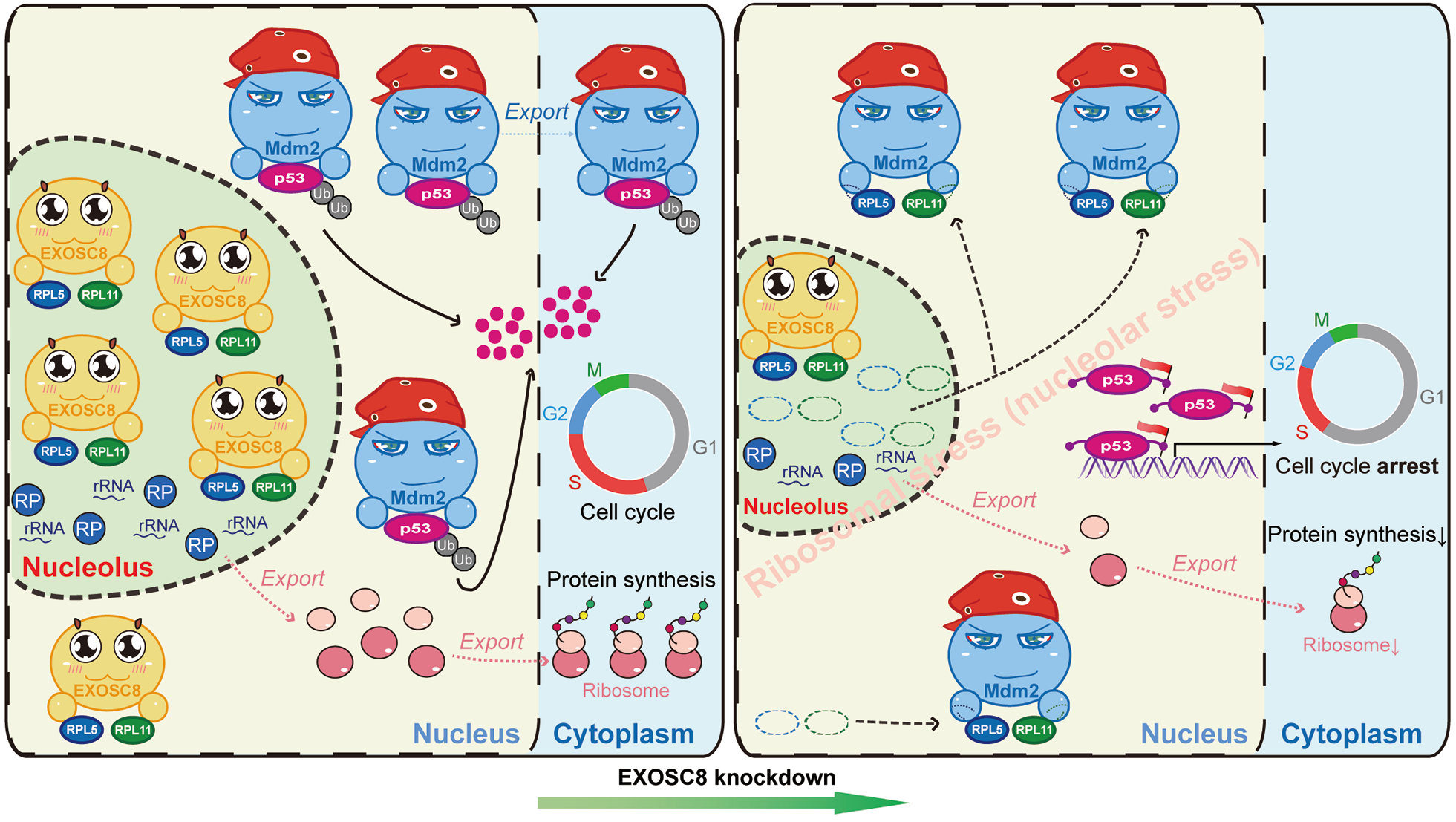
Certainly, we have noted that our study has some limitations due to the influence of COVID-19 and our conditions. For instance, more assays and wet-lab experiments could be performed to validate the relationship among EXOSC8, ribosome biogenesis-related processes, RP5/11-Mdm2-p53, ribosomal stress and tumorigenesis; More mice could be used in therapy experiments. Nonetheless, our study highlights the significance of EXOSC8 in CRC. EXOSC8 is upregulation driven by CNV gain and correlated with poor prognosis in CRC. For the first time, the RNA exosome gene (EXOSC8) was confirmed to play a critical role in cancer-related ribosome biogenesis and RPs-Mdm2-p53-mediated ribosome biogenesis surveillance pathway in cancer cells. This work further extends the previous Pan-cancer study of the rRNA metabolism-related genes [1]. Importantly, inhibiting oncogene-driven upregulation of ribosome biogenesis in human cancer provides therapeutic specificity for selectively targeting cancer cells rather than normal cells [14]. Hence, our findings suggest that inhibition of EXOSC8 is a novel therapeutic strategy for targeting ribosome biogenesis and p53 signaling in CRC.
In the future, we will identify new rRNA metabolism-related genes and explore their functions and mechanisms in human cancers. Furthermore, I and my co-authors continuing a series of Pan-cancer studies on TME, tumor metabolism and programmed cell death, etc.
1 Cui K, Liu C, Li X, Zhang Q, Li Y. Comprehensive characterization of the rRNA metabolism-related genes in human cancer. Oncogene. 2020; 39: 786-800.
2 Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45: 1113-1120.
3 Boczonadi V, Muller JS, Pyle A, Munkley J, Dor T, Quartararo J et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat Commun. 2014; 5: 4287.
4 Zanchin NI, Goldfarb DS. The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res. 1999; 27: 1283-1288.
5 Cui K, Yao S, Zhang H, Zhou M, Liu B, Cao Y et al. Identification of an immune overdrive high-risk subpopulation with aberrant expression of FOXP3 and CTLA4 in colorectal cancer. Oncogene. 2021; 40: 2130-2145.
6 Cui K, Yao S, Liu B, Sun S, Gong L, Li Q et al. A novel high-risk subpopulation identified by CTSL and ZBTB7B in gastric cancer. Br J Cancer. 2022; 127: 1450-1460.
7 Cui K, Gong L, Wang K, Wang Y, Huang L, Liu B et al. Ferroptosis-Associated Molecular Features to Aid Patient Clinical Prognosis and Therapy Across Human Cancers. Front Immunol. 2022; 13: 888757.
8 Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018; 18: 51-63.
9 Muller JS, Burns DT, Griffin H, Wells GR, Zendah RA, Munro B et al. RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels. Life Sci Alliance. 2020; 3.
10 Shen A, Chen Y, Liu L, Huang Y, Chen H, Qi F et al. EBF1-Mediated Upregulation of Ribosome Assembly Factor PNO1 Contributes to Cancer Progression by Negatively Regulating the p53 Signaling Pathway. Cancer Res. 2019; 79: 2257-2270.
11 Cao P, Yang A, Li P, Xia X, Han Y, Zhou G et al. Genomic gain of RRS1 promotes hepatocellular carcinoma through reducing the RPL11-MDM2-p53 signaling. Sci Adv. 2021; 7.
12 Li D, Yao S, Zhou Z, Shi J, Huang Z, Wu Z. Hyaluronan decoration of milk exosomes directs tumor-specific delivery of doxorubicin. Carbohydr Res. 2020; 493: 108032.
13 Gong L, Li Y, Cui K, Chen Y, Hong H, Li J et al. Nanobody-Engineered Natural Killer Cell Conjugates for Solid Tumor Adoptive Immunotherapy. Small. 2021; 17: e2103463.
14 Kang J, Brajanovski N, Chan KT, Xuan J, Pearson RB, Sanij E. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther. 2021; 6: 323.
Follow the Topic
-
Oncogene

This journal aims to make substantial advances in our knowledge of processes that contribute to cancer by publishing outstanding research.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in