New insights into CTSL/ZBTB7B in gastric cancer prognosis and tumor microenvironment
Published in Cancer

Gastrointestinal (GI) cancer is one of the solid cancer types with a complex and heterogeneous tumor microenvironment (TME) 1,2. Last year, we have reported a unique subpopulation of regulatory T cells with intense CTLA4 expression in colorectal cancer (CRC) 3. This subgroup showed a high-risk of death and recurrence with an immune overdrive TME phenotype, including high levels of immune infiltration, immune checkpoints (ICs) expression, MSI/dMMR status and TGF-β signaling. After that, Prof. Huang and Prof. Fei ask me, as one of the prevalent type of GI cancer, whether gastric cancer (GC) showed similar phenotypes based on two markers. Thus, we firstly used the method publishing in our previous study to analyze ICs survival in GC 3. However, most ICs were associated with a favorable OS or did not show an obvious relationship with survival, let alone double peaks with significant P values like our previous study in CRC 3.

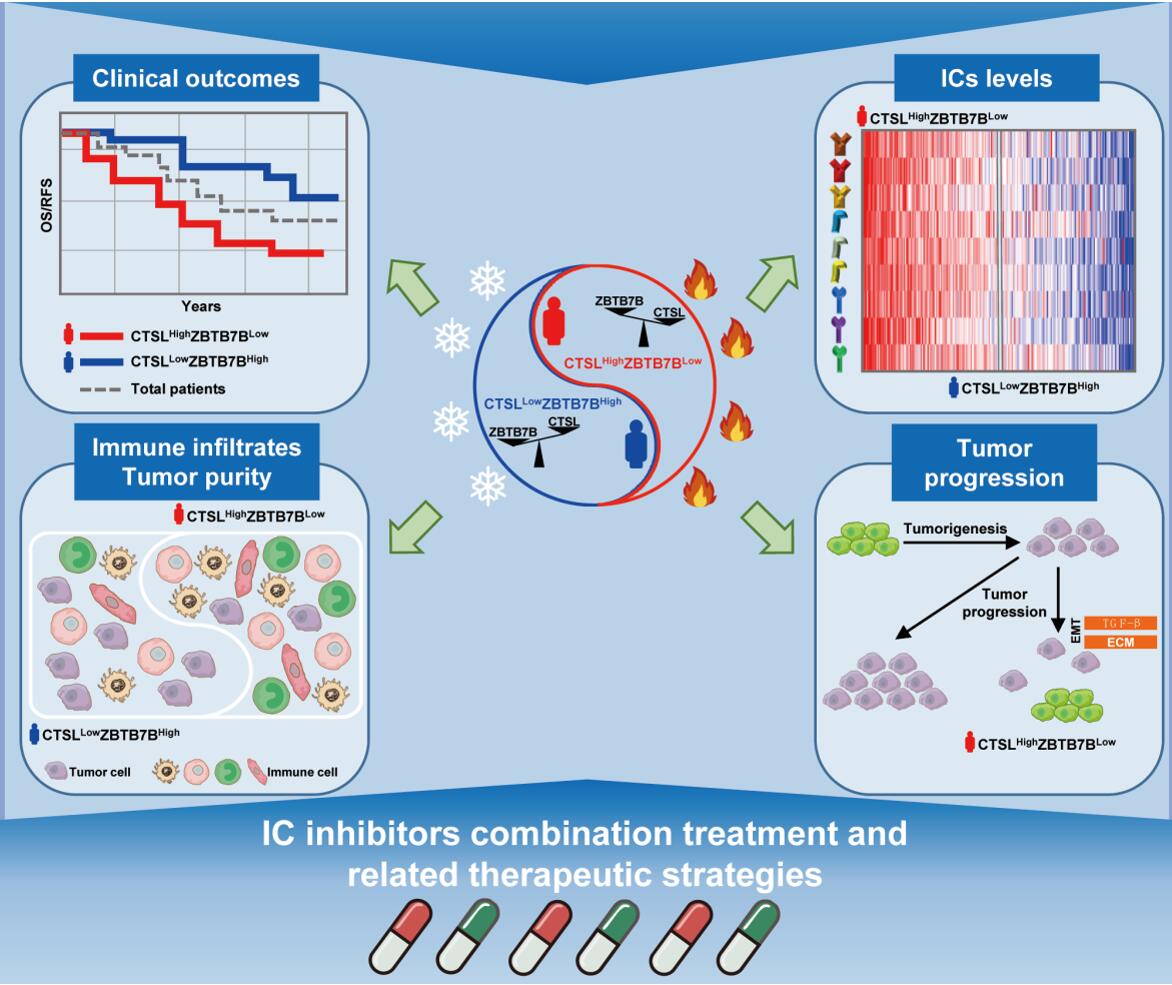
Therefore, we designed another identification approach to find ICs-related survival predictors in GC. Subsequently, CTSL/ZBTB7B is identified as the top ICs-related gene that represents poor/favorable clinical. CTSL is a human cysteine proteases gene, and ZBTB7B is a CD4-lineage transcription factor gene. We observed that CTSL was upregulated and positively correlated with poor survival and ICs expression, while ZBTB7B showed contrary results. Survival prediction among the combination of CTSL/ZBTB7B was better than single CTSL or ZBTB7B according to P values. Moreover, the stratification based on the expression levels of CTSL and ZBTB7B were intensely correlated with survival/recurrence and remained an independent prognostic variable for GC patients based on multiple cohorts.
Next, we performed the TME analysis and observed that the CTSLHighZBTB7BLow high-risk subpopulation featured high immune infiltrates and low tumor purity. This result makes us a little confused because CTSL seems as a tumor-promoting gene, while ZBTB7B as a tumor-suppressing gene according to the abovementioned expression/survival analyses and previous reports in GC 4,5. Exhilaratingly, the availability of single-cell transcriptomic profiles across a broad range of human cancers provides an unprecedented opportunity to explore TME in greater depth. Emerging evidence showed that GC malignant cell population expressed relatively higher ZBTB7B, whereas CTSL expression levels were higher in nonmalignant cell types, especially in macrophages. Intriguingly, before submitting this manuscript to BJC, we found a newly published study about TME that identified tumor-associated macrophages (TAM) derived from bone marrow or purified from E0771 tumors overexpressed CTSL and harbored hyperactive cysteine protease activity in their lysosomes 6. Incubating the antigenic peptide gp10025–33 with CTSL before delivering it to TAMs blocked their ability to activate CD8 T cells according to CD8 T cells and TAMs co-culture assay. This study further enhanced our confidence in our research of CTSL. We performed multicolor immunofluorescence staining in GC samples according to reviews’ comments and confirmed that CTSL was co-expressed to macrophage marker CD68.
In this study, we uncovered a poor survival-related subgroup CTSLHighZBTB7BLow but featured high immune infiltrates, low tumor purity and high ICs levels, suggesting an immune overdrive phenotype between immunostimulatory and immunosuppressive mechanisms. Noted that this subgroup is dissimilar to previous immune overdrive TME phenotypes, such as high intratumoral CD8 T cell infiltration with a high density of TAMs that expressed CD274, or Tregs with overexpressed CTLA4 in CRC 3,7. Moreover, traditional cancer studies generally used cancer cell lines to explore the tumor-promoting/suppressing function. In this study, our results suggest that some tumor-promoting genes affect tumorigenesis and progression by regulating the functions of nonmalignant cell populations. For instance, CTSL as a tumor-promoting gene that is mainly expressed in nonmalignant cells, whereas ZBTB7B as a tumor-suppressing gene that is mainly expressed in malignant cell type. We also reported that CD274 is lower expressed in malignant cells but highly abundant in TAMs, and a specific CD274+CD206+ macrophage subpopulation was associated with poor survival in CRC 8. These studies suggest that TME should be further considered in the future work of tumor-promoting/suppressing function. Certainly, we have noted that the biological relationship between CTSL and ZBTB7B for poor prognosis in GC is yet to be determined and likely further require dynamic single-cell analysis of immune contexture in the future. Besides, as discussed in a previous TME study, the single-cell transcriptomic profiles and immunofluorescence staining normally assumed all-cell populations summed up as 100% in current conditions 9. In the real TME, the number of immune cells may be increased or reduced with different percentages.
In conclusion, we established a CTSL/ZBTB7B subtyping system, and uncovered the novel CTSLHighZBTB7BLow high-risk subgroup, but characterized by relative higher immune cell infiltration and lower tumor purity. This subgroup demonstrates higher levels of ICs and more enrichment of cancer-related pathways compared with other cases. This study further provides a personalized prognostic method and may contribute to precision cancer therapies. Patients from the CTSLHighZBTB7BLow subpopulation may benefit from more active IC inhibitor-based combination treatment, which will also help in the development of related therapeutic strategies.
References
1 Punt CJ, Koopman M & Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235-246.
2 Ramezankhani R, Solhi R, Es HA, Vosough M & Hassan M. Novel molecular targets in gastric adenocarcinoma. Pharmacol Ther. 2021;220:107714.
3 Cui K, Yao S, Zhang H, Zhou M, Liu B, Cao Y, et al. Identification of an immune overdrive high-risk subpopulation with aberrant expression of FOXP3 and CTLA4 in colorectal cancer. Oncogene. 2021;40:2130-2145.
4 Pan T, Jin Z, Yu Z, Wu X, Chang X, Fan Z, et al. Cathepsin L promotes angiogenesis by regulating the CDP/Cux/VEGF-D pathway in human gastric cancer. Gastric Cancer. 2020;23:974-987.
5 Xia L, Jiang L, Chen Y, Zhang G & Chen L. ThPOK transcriptionally inactivates TNFRSF12A to increase the proliferation of T cells with the involvement of the NF-kB pathway. Cytokine. 2021;148:155658.
6 Cui C, Chakraborty K, Tang XA, Schoenfelt KQ, Hoffman A, Blank A, et al. A lysosome-targeted DNA nanodevice selectively targets macrophages to attenuate tumours. Nat Nanotechnol. 2021;16:1394-1402.
7 Fakih M, Ouyang C, Wang C, Tu TY, Gozo MC, Cho M, et al. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J Clin Invest. 2019;129:4464-4476.
8 Yin Y, Liu B, Cao Y, Yao S, Liu Y, Jin G, et al. Colorectal Cancer-Derived Small Extracellular Vesicles Promote Tumor Immune Evasion by Upregulating PD-L1 Expression in Tumor-Associated Macrophages. Adv Sci (Weinh). 2022;9:2102620.
9 Ye Y, Zhang Y, Yang N, Gao Q, Ding X, Kuang X, et al. Profiling of immune features to predict immunotherapy efficacy. Innovation (Camb). 2022;3:100194.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in