The Story Behind the Paper: From a Workshop to a Published Fight Against Malaria
Published in Chemistry
Two years ago, I attended a molecular docking workshop, eager to learn how virtual screening could accelerate the discovery of new drugs. Little did I know that a simple suggestion from the coordinator—"Why don’t you all form a group and continue a project after this?"—would set us on a path leading to a peer-reviewed publication in Discover Chemistry.
Our newly formed team decided to tackle a problem close to home: malaria. In Nigeria, malaria accounts for a staggering burden—30% of childhood deaths, 11% of maternal deaths, and over 60% of clinical visits. With the terrifying rise of artemisinin-resistant Plasmodium falciparum strains, the need for new therapeutic strategies has never been more urgent.
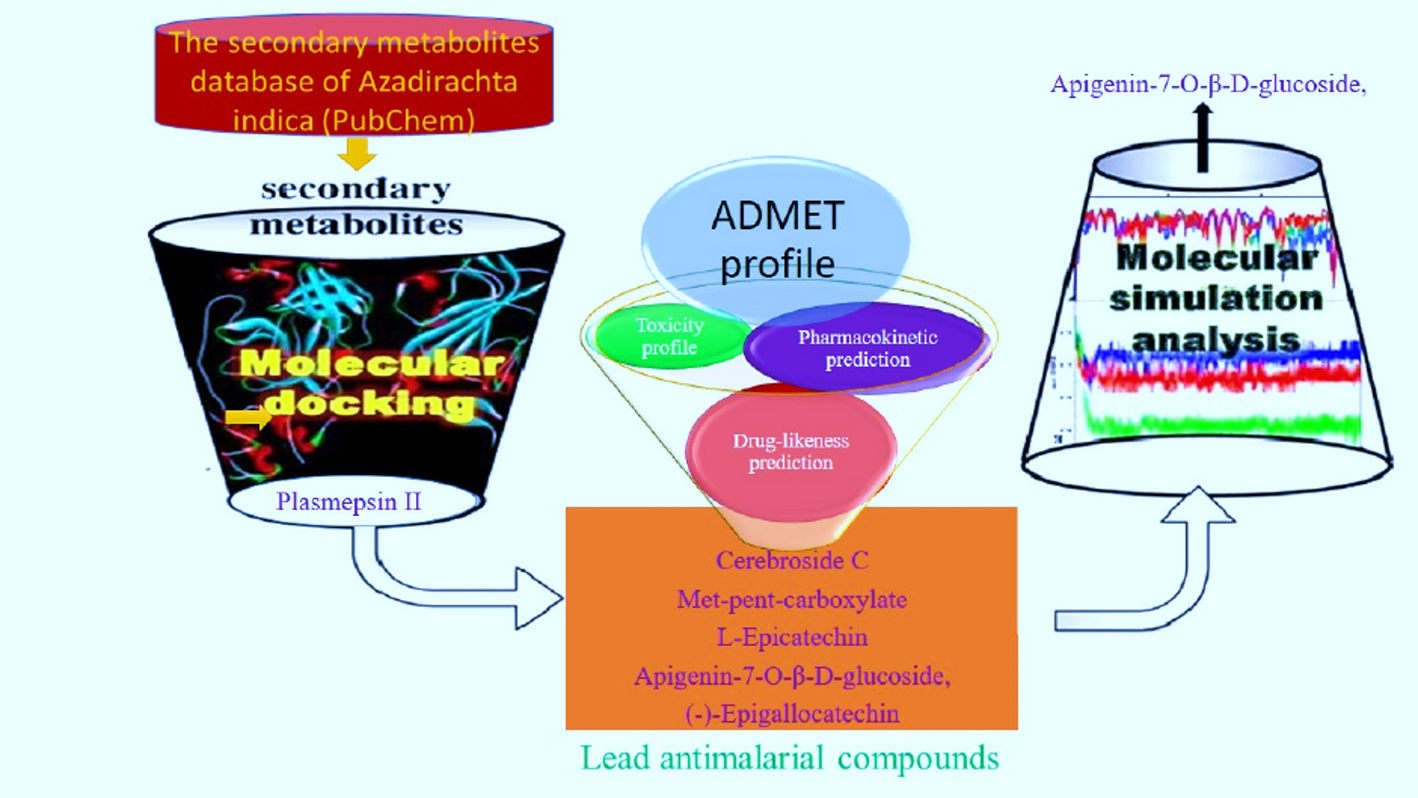
We chose to look for answers in a familiar guardian: the Neem tree (Azadirachta indica). While its traditional uses are well-known, its vast chemical repertoire remained largely unmapped for modern, target-based drug discovery. Our mission was clear: use computational tools to systematically mine neem's 320+ phytochemicals for a specific weapon—an inhibitor of Plasmepsin II, a critical enzyme the malaria parasite uses to digest hemoglobin and survive in our blood.
As the group leader, I witnessed an incredible synergy. We pooled our resources to present initial findings at conferences. Then began the real grind: nights of running docking simulations, analyzing complex pharmacophore models, and tracking the dynamic dance of protein-ligand interactions across 100 nanoseconds of simulation time.
The results were illuminating. Our screen identified Cerebroside C as a potent binder, with the highest predicted affinity. A classic approach might have stopped there. But our deeper analysis revealed a crucial lesson in drug discovery: binding strength alone is not enough.
Through ADMET profiling, we found Cerebroside C would likely have poor bioavailability and be pumped out of cells by P-glycoprotein. Instead, a compound named Apigenin-7-O-β-D-glucoside emerged as the dark horse. It showed strong, stable binding in dynamic simulations, forming a resilient complex with key residues like Asp214 and Tyr77, and—most importantly—it possessed the drug-like properties necessary to potentially become a real medicine.
This research, now published, is more than just a paper. It is a testament to collaborative spirit, from funding our own initial steps to supporting each other through the manuscript process. It highlights the power of computational foresight—using in silico tools to guide the search for therapeutics, saving time and resources. Most importantly, it adds a promising piece to the global puzzle of defeating drug-resistant malaria.
My deepest thanks go to my incredible co-authors and teammates: Garba Dauda, Amina Busola Olorukooba, Iyabo Mobolawa Adebisi, Omobolanie Ibukun Ogundele, Murtala Muhammad Jibril, Aiseosa Kingsley Iqhar, Jamilu Sani, Walter Mdekera Iorjim, Okesola Mary Abiola, Abayomi Emmanuel Adegboyega, and Titilayo Omolara Johnson. This achievement is yours as much as it is mine.
The journey from a workshop idea to a published study reminds us that scientific progress often starts with a simple step: a curious team deciding to work together on a problem that matters.
Read our full open-access story here: https://rdcu.be/eXrW8
DOI: 10.1007/s44371-025-00461-z
#ScienceStory #MalariaResearch #DrugDiscovery #Neem #ComputationalBiology #TeamScience #NigerianResearch #AcademicTwitter #FromWorkshopToPublication
Follow the Topic
-
Discover Chemistry

A fully open access, peer-reviewed journal supporting multidisciplinary research and policy developments across all fields of chemistry.
Related Collections
With Collections, you can get published faster and increase your visibility.
Material Chemistry in Biomedical Applications
Chemistry-driven processes are essential inspirations for researchers to design biomaterials applied as promising diagnostic tools, therapeutic solutions or tissue substitutes, etc. To increase the understanding of material chemistry in biomedical applications, this topical collection focuses on documenting the chemistry covering nanoscale to macroscale biomaterials, including surface science and interactions with the body at the molecular level. We seek articles that highlight the principles of material chemistry or approaches to investigate and improve the material properties in biomedical research. The material properties here mainly include biocompatibility, physical, chemical, mechanical, and other particular properties required for medical and dental materials. Reviews, original research, and commentaries are all welcomed.
Keywords: Material chemistry, surface science, nanotechnology, biocompatibility, physical and chemical property, mechanical property, biomedical application.
Publishing Model: Open Access
Deadline: Mar 31, 2026
Advances in Material Chemistry for Life Science Research
Material chemistry has a long-standing history and has been extensively exploited in a wide range of life science research such as biological, chemical, and environmental engineering. The invention of novel materials for biological applications has led to the creation of biomaterials for tissue engineering, nanomaterials for drug delivery, and smart materials that interact dynamically with biological systems. These advances have significantly revolutionized the fields of healthcare including medical treatments, diagnostics, and biological studies. In recent years, the synergy between material chemistry and life sciences has been particularly evident in the development of materials that enable precision medicine, improve the accuracy of diagnosis, and facilitate regeneration of defected tissues. In particular, nanoparticles engineered for targeted drug delivery have significantly enhanced the efficacy of treatments while minimizing side effects, and responsive biomaterials have opened new avenues for developing adaptive therapeutic devices. The application of these advanced materials in life science not only enhances our understanding of biological processes but also paves the way for innovative solutions to pressing healthcare challenges. This topical collection aims to cover papers that explore the intersection of material chemistry and life sciences, focusing on the design, synthesis, and application of advanced materials in biological contexts. Translational advances in material chemistry for healthcare, biotechnology, and environmental sustainability will be highlighted in this topical collection, reflecting the multidisciplinary nature of this rapidly evolving field.
Keywords: Material chemistry, life science, biomaterials, smart materials, drug delivery, nanomaterials, medicinal chemistry.
Publishing Model: Open Access
Deadline: Jun 30, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in