Uncovering diversity and metabolic spectrum of animals in dead zone sediments
Published in Ecology & Evolution

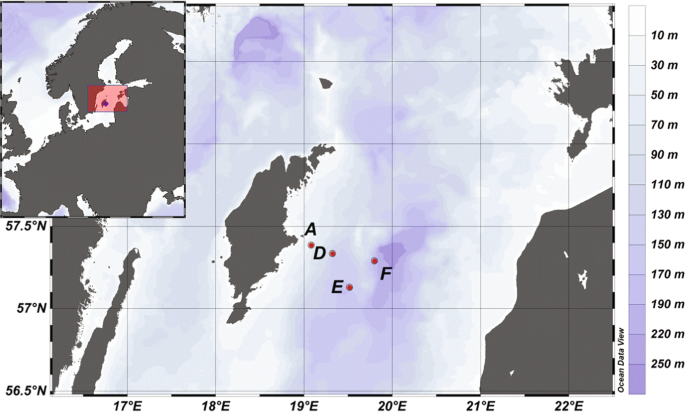
We have conducted several cruises during the last few years to this Baltic Sea dead zone, and for this study the data comes from sampling during April 2018. This is a month that generally presents surprises when you are in the middle of the Baltic, as one day it can be the beginning of the spring with even the visit of passerine birds in the middle of the sea. And the day after it can be close to full-winter conditions with thick clouds bringing some snow and rough sea conditions making the ship (R/V Skagerak from the University of Gothenburg) rocking.

During sampling we managed to get water and sediment samples from all the four stations we visited. Sediment was collected for both i) microprofiling of geochemical characteristic and ii) slicing the surface layer that was flash frozen for later RNA extraction. From the environmental point of view, dead zones are among one of the most inhospitable places on Earth. Many areas are completely devoid of oxygen, and if oxygen is present it is gone already at a few millimeters under the sediment surface. In the absence of oxygen, the poisonous gas hydrogen sulfide is produced by bacteria in the sediment, and already at low concentrations this gas is toxic to most animals because it inhibits vital biochemical reactions happening in cell’s mitochondria.

In the lab we extracted RNA from the sliced sediment samples and sequenced it. During the bioinformatic analyses of the RNA data, we extracted 18S rRNA sequences and found a large diversity of single-cell eukaryotes (protists) and fungi. Many of these small organisms are known to survive by anaerobic metabolism and it was not surprising that they dominated our dataset. Also as expected, we couldn't find molecular signals of macrofauna (invertebrates larger than 1 mm) and other macroscopic animals. But we did find these clues for smaller animals. Arthropoda and Rotifera were present at all stations which made us suspect that these are likely resting eggs from zooplankton. Moreover, we noticed that the relative abundance of 18S rRNA sequences for nematodes among the samples showed a different pattern compared to the other animals. They were present in all dead zone samples, and remarkably different in diversity and taxonomy, depending on the geochemical conditions for each station. We confirmed this discovery by analyzing the mRNA data, and were a bit surprised how well the pattern of nematode mRNA transcripts matched the pattern of the nematode 18S rRNA sequences. These nematodes seemed to be there and alive according to the molecular data. The next step was to check that we could see these nematodes in the same sediments with our eyes, which was confirmed via microscopy.
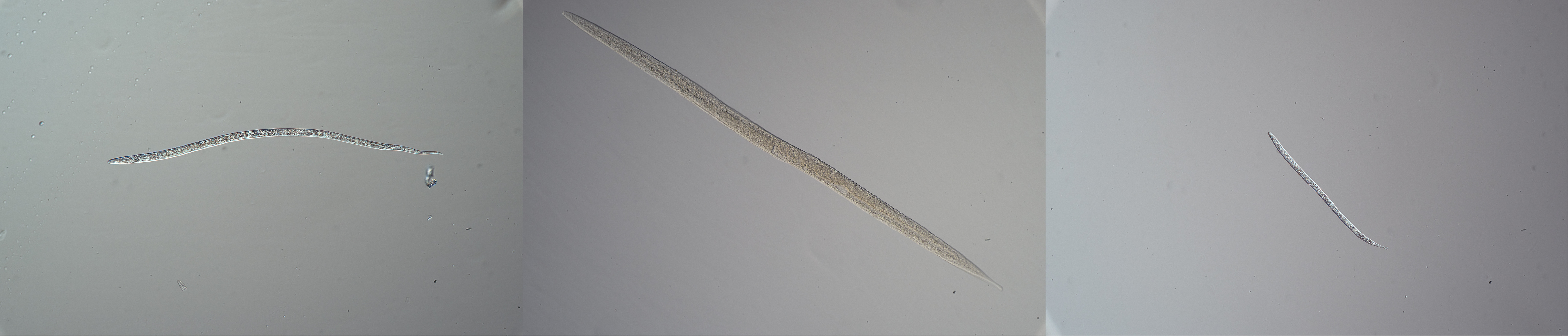
To conclude, in this study by Broman, Bonaglia et al. we were able to combine a large dataset of both geochemical data and molecular data. We show the presence of potentially alive animals in dead zone sediments with the use of microscopy and RNA sequencing, with results from both 18S rRNA and RNA transcripts.
The published article is available at:
https://www.nature.com/articles/s42003-020-0822-7
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in