A genetic strategy to combat antibiotic resistance that can spread between bacteria
Published in Bioengineering & Biotechnology
Background
Bacterial antibiotic resistance (AR) poses a serious global health threat and is currently responsible for approximately 1.27 million deaths worldwide, a toll that is estimated to soar to over 10 million deaths per year by 2050 if left unchecked. Important factors contributing to the increase in AR include the widespread over-prescription of antibiotics, their misuse in animal husbandry, inadequate sewage treatment, and environmental contamination. These troubling trends have converged to spawn the emergence and spread of multidrug-resistant bacteria or “superbugs” that could overwhelm current antimicrobial interventions. Thus, there is a pressing need for discovering new antibiotics and innovating complementary strategies to combat AR.
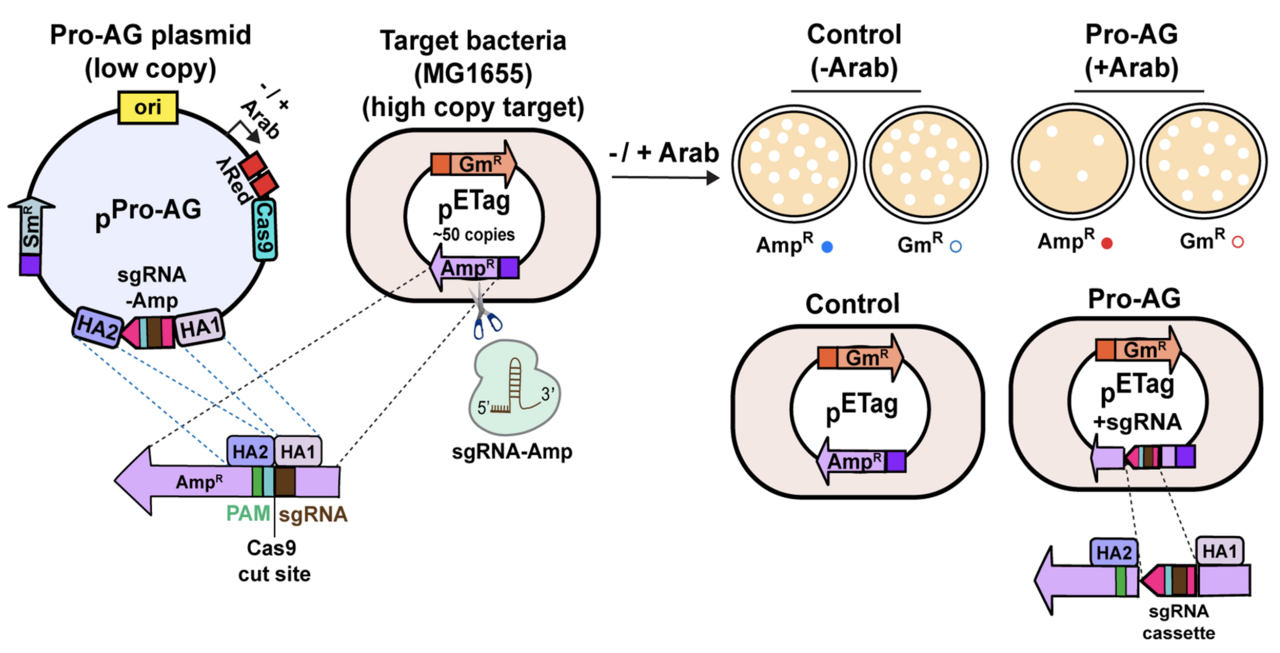
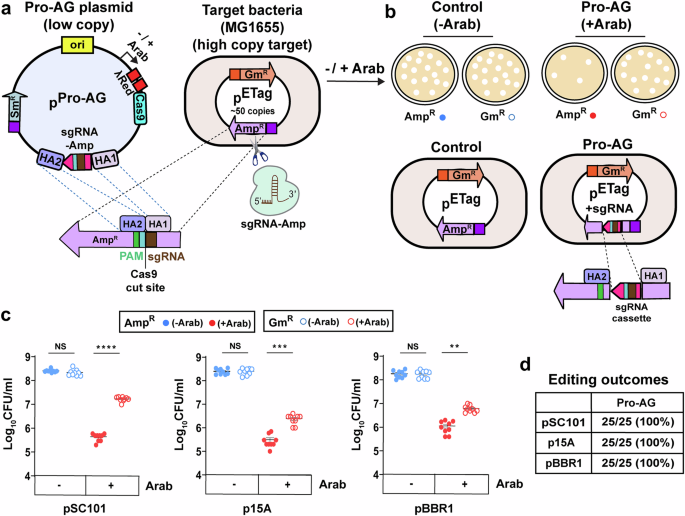
One strategy, dubbed Pro-Active Genetics (Pro-AG), was developed previously in a close collaborative effort between my group and Victor Nizet's lab at UC San Diego School of Medicine) (Valderrama et al., 2019; Nat. Comm. 10, 5726). This CRISPR-based technology is similar to the methodology underlying so-called gene-drives in insects (also developed in the Bier lab - see Bier, 2022; Nat. Rev. Genet. 23, 5-22) wherein a genetic cassette is efficiently copied between genomes. In case of Pro-AG, this copying takes place between two autonomously replicating plasmids, which often carry genes conferring antibiotic resistance. The prototype Pro-AG system we developed using multiple plasmids targeted insertion of a gene cassette carrying a single guide RNA (sgRNA) into the b-lactamase gene to inactivate it and thereby restore sensitivity to the antibiotic Ampicillin. Compared to prior CRISPR schemes designed to cut and destroy AR bearing plasmids, which resulted in ~100 fold reduction in AR, the Pro-AG system reduced the frequency of AR colonies by ~100,000 fold. A key factor underlying efficiency of Pro-AG was a positive feedback cycle wherein copying of the sgRNA bearing cassette into the target plasmid increased levels of the sgRNA, which proved to be a limiting component.
Current Study
In a new collaborative study together with the Meyer lab (University of California, San Diego), first author Saluja Kaduwal (Bier lab) developed supped up "Pro-AG-MobV" platform that combines the separate CRISPR components (e.g., Cas9, sgRNA, and lambda-Red recombination cassette) from the original prototype system (Kaduwal et al., 2026; npj Antimicrob. Resist. 4, 8). In addition to consolidating these elements into a unitary systems the Pro-AG-MobV plasmid is also endowed with ability to spread between bacterial cells by a process known as conjugal transfer - the mating equivalent for bacteria (See Figure). We also characterized another highly efficient process that eliminates the AR gene, referred to as homology based deletion (HBD), wherein a target gene flanked by short repeated sequences can be deleted by directing Cas9 cutting to anywhere between the repeats. Saluja analyzed the mechanistic underpinnings of HBD, and applied it to creating a mitigating system for deleting a potentially undesired gene cassette (e.g., one carrying Pro-AG components) as a safety measure akin to technologies that have developed to eliminate errant gene-drives in insects (Xu et al., 2020; Molecular Cell 80, 246-262) should the need arise.
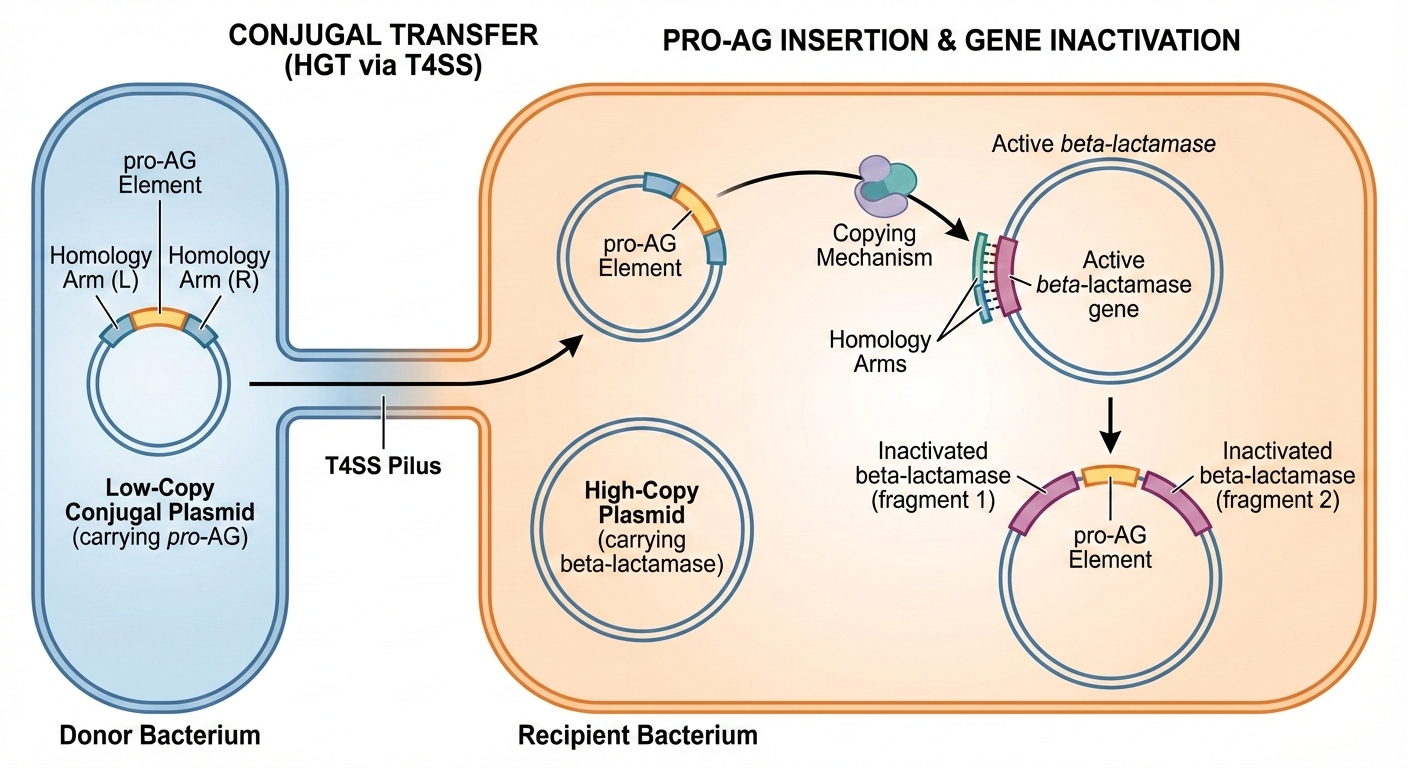
Saluja also showed that the Pro-AG-MobV plasmid is indeed capable of spreading between bacterial cells as intended, and moreover that it does so with particular efficiency when bacteria coat surfaces in biofilms. This context for combatting AR is particularly important since biofilms are one of the most challenging forms of bacterial growth to combat in the clinic or in enclosed environments such as aquafarm ponds or sewage treatment plants.
Although we focus in this study on showing that the new all-in-one Pro-AG-MobV system can efficiently reduce AR in bacterial population by as much as five orders of magnitude, we point out that the same technology should be readily adaptable for a broad variety of purposes to efficiently edit bacterial genes responsible for pathogenic bacterial behaviors. In addition, Saluja and Elizabeth Stuart from the Meyer lab show that components of the Pro-AG system also can be carried by a bacterial virus (bacteriophage or phage). This dual plasmid/phage carriage opens the door to engineering new versions of Pro-AG plasmids and viruses that can act in a coordinated fashion to counter the evolution of bacterial resistance to phage treatments.
Follow the Topic
-
npj Antimicrobials and Resistance

This journal considers basic, applied, and clinical research that advances our understanding of all aspects of antimicrobials and antimicrobial resistance.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data Science, AI and Machine Learning Approaches to AMR
Publishing Model: Open Access
Deadline: Apr 07, 2026
Antimicrobials and Resistance in the Food Chain
Publishing Model: Open Access
Deadline: Jul 08, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in