A scarless guide to modulating gene expression
Published in Bioengineering & Biotechnology

Background context: Development of the two-component Cas9/guide-RNA (gRNA) CRISPR system for precise genome editing by Jennifer Doudna and colleagues is one of the major breakthroughs of this century, and earned the 2020 Nobel Prize in Chemistry. Specific DNA cleavage can be achieved in vivo with remarkable efficiency using gRNAs designed with 20 nucleotide (nt) complementarity to the genome target. Since then, a myriad of alternative CRISPR uses have been developed by altering the activity of the Cas9 protein. Several of these approaches make use of a Cas9 variant in which both DNA-nicking catalytic centers are mutated to create an enzymatically dead form dubbed dCas9. A variety of other protein domains can be fused to either the N- or C-terminus of Cas9 to generate new sequence-specific activities. For example, by fusing transcriptional repression domains to dCas9 coupled with a library of gRNAs targeting binding to regions near transcriptional start sites of genes, it is possible to perform genome-wide CRISPR-inhibition (CRISPRi) screens to test the effects of gene knock-down on various cellular phenotypes such as growth or resistance to toxins. Reciprocally, one can activate target genes by fusing a transcriptional activation domain to dCas9 for CRISPR-activation (CRISPRi-a) screens. In addition, one can fuse proteins with other functionalities to carry out site-specific base-editing, chromatin alteration, or visualization of nuclear processes.
Studies of the molecular activity of Cas9 have revealed a sequence of structural changes and catalytic "checkpoints" regulating the ability of Cas9/gRNA complexes to recognize, bind and ultimately cleave specific DNA targets. The multi-step nature of the final event permits a separation of sequence-specific DNA binding from subsequent double strand cleavage. For example, when combined with a truncated gRNA (tgRNA) shorter than the standard 20 nt guide (i.e., <16nt), Cas9 can bind, but not cut target sequences, since pairing of the last 4 nucleotides of gRNA with target are required to trigger this final event. Thus, high levels of binding specificity are retained while cleavage is abrogated.
Over the past several years, my research group has helped develop CRISPR-based "active genetic" elements referred to as gene-drives that encode Cas9 and a gRNA to direct Cas9/gRNA cleavage at the precise site where the gene cassette is inserted into the genome. Such gene-drive elements can be transmitted to progeny in a super-Mendelian fashion (>50%) as a result of the action of the homology directed repair (HDR) pathway and gene conversion of the homologous chromosome in the germline. Highly efficient gene-drives have been developed in insects such as fruit flies (Drosophila), Anopheline mosquitos as well as single-celled yeast and bacteria (e.g., ~99% transmission), and similar systems have also shown promise in other species including mammals (mice).
In addition to carrying Cas9 and a gRNA for copying itself, a gene-drive cassette can also include "cargo" with beneficial activities such as expression of single chain antibodies in the midguts of blood-feeding female mosquitoes that block transmission of malarial parasites. Auxiliary gRNAs could also be included to a gene-drive to target cleavage of undesirable allelic variants of relevant target genes for replacement by favored variants. For example, variants of the voltage-gated sodium ion channel conferring resistance to insecticides, or specific variants of host mosquito factors that aid malarial parasite transmission (such as the intestinal protein FREP1) could be progressively eliminated in natural populations using such accessory gRNAs. A concern, however, is that such strategies deploying multiple gRNAs may result in multiple genomic cleavage events (i.e., at the site of the gene-drive insertion and at the secondary targets), which could lead to chromosome rearrangements or genomic damage with potentially undesired consequences.
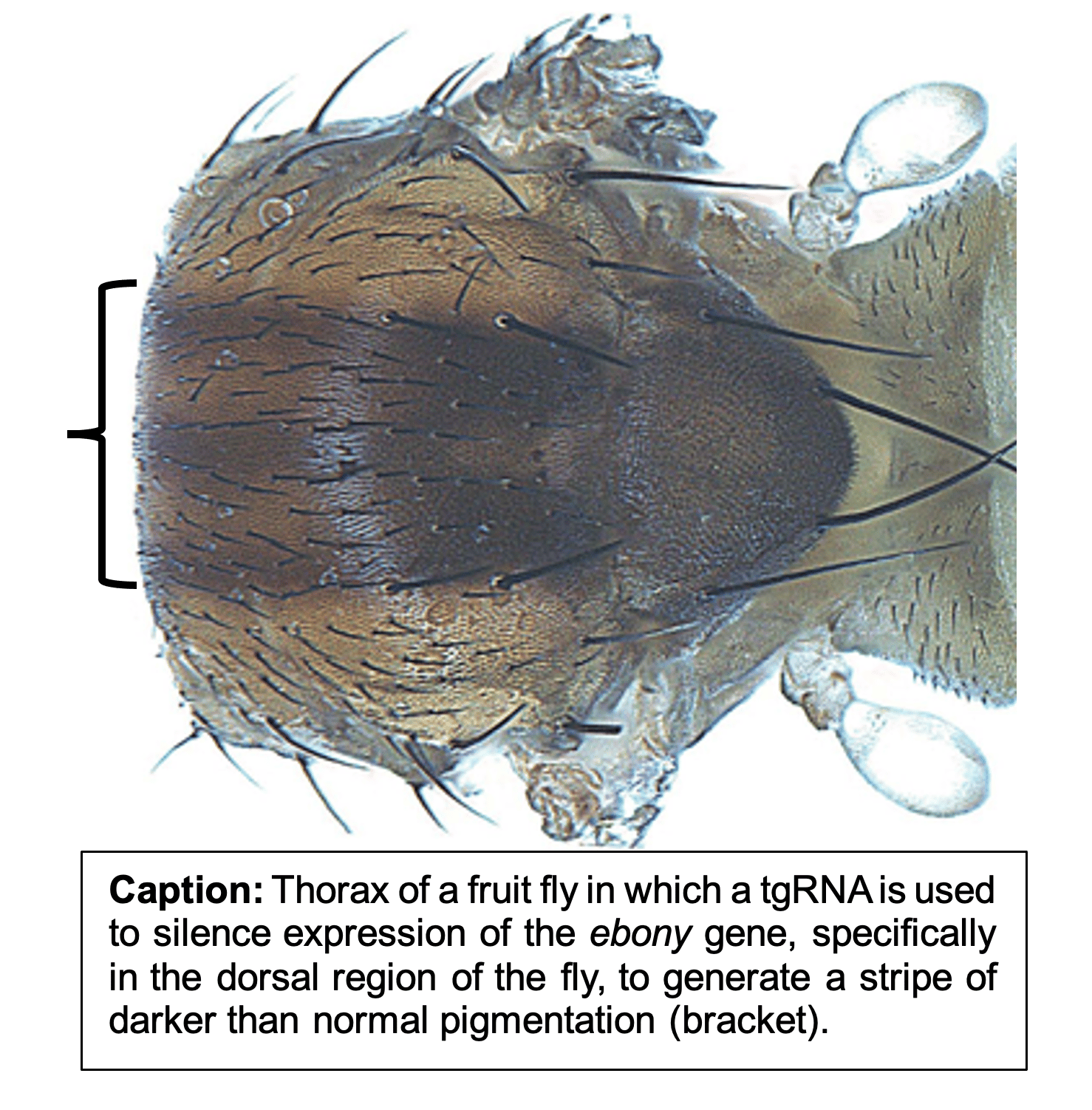
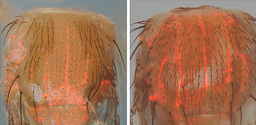
Current Study: Now, in a newly published study, Ankush Auradkar and colleagues devise a hybrid gene-drive system based on a combination of full-length and truncated gRNAs. In such drives, the active form of Cas9 is used to sustain both copying of the drive element (using the full-length gRNA) and modulation of gene expression by employing tgRNAs to either repress (tgCRISPRi) or activate (tgCRISPRa) transcription of secondary target genes without causing any genomic damage. Auradkar et al. show that tgRNAs targeting sites between the transcription start site (TSS) or upstream TATA boxes can lead to efficient gene knock-down (>80% reduction). This strategy can operate successfully for genes including a TATA box or not (TATA-less promoters). Also, fusing a transcriptional activation domain to the otherwise active Cas9 protein (Cas9-VPR) can lead to overexpression or mis-expression of the target gene. Furthermore, this scarless gene regulatory approach can be extended to target distant cis-acting regulatory elements (referred as enhancers or cis-regulatory modules = CRMs) of otherwise essential genes (i.e., genes whose function is required for organismal viability or fertility). Such CRM-targeted tgRNAs can either repress (Cas9) or activate (Cas9-VPR) target gene expression in limited regions of the organisms while preserving viability. For example, tgRNAs targeting a CRM of the HOX segment identity gene Sex Combs Reduced (Scr), which is required specifically for formation of male mating bristles, leads to a reduced number of such bristles. Likewise, tgRNAs targeting a repressor binding site in a CRM of the essential gene knirps, (which directs specific expression in the second longitudinal wing vein), leads to an anterior displacement of this vein in the presence of Cas9 due to reduced repression, while generating ectopic veins in the presence of the activating Cas9-VRP source.
Auradkar et al. show further that tgRNAs and traditional full-length gRNA can be used concomitantly to effectively silence a target gene (tgRNA) and sustain copying of the gene-drive (full length gRNA). This observation opens up the possibility of using tgRNAs to improve gene-drive copying efficiencies in certain systems either by silencing genes that reduce copying (i.e., genes involved in error-prone non-homologous end-joining repair) or by promoting expression of genes in the preferred HDR repair pathway (e.g., using Cas9-VRP). Additionally, tgRNAs might also be added to gene-drives in mosquitos to reduce transmission of disease pathogens such as malarial parasites or various viruses. In the case of malaria, such tgRNAs could either silence expression of host mosquito genes required for parasite development or enhance expression of innate immune genes to eliminate the parasites.
The work continues: Applications of the technology developed in the Auradkar study also extends beyond the field of gene-drives. For example, co-author Saluja Kaduwal showed that a similar tgRNA system could be deployed in human cells to reduce expression of a target gene. These applications are particularly exciting to co-authors Annabel Guichard and Ketta Sneider who have been developing CRISPR systems for scarless copying of preferred allelic variants in somatic cells of the body using so-called Nickase versions of Cas9 in which only one of the two catalytic centers of the enzyme has been mutated. The use of dual acting gRNAs for copying a gene cassette or preferred allelic copying (full length gRNA) and for altering expression of other genes (tgRNAs) that could aid in potential gene-therapy outcomes (e.g., immune modulatory functions) has broad applications. These are the exciting new directions that our group, and hopefully many others, will take using tgRNA-inclusive gene-editing systems.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in