A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules
Published in Chemistry

Ligand discovery is one of the key steps in biomedical research for applications ranging from drug discovery to diagnostic tools. Identifying ligands that target protein-protein interactions is more challenging because of their large contact area, hydrophobic nature, and flat surface characteristics, making them unsuitable to be targeted by small molecules. These factors often categorize the protein-protein interactions under the ‘undruggable’ domain. Despite such challenges, nature has evolved an elegant way of targeting protein-protein interactions via antibodies. Antibodies inhibit protein-protein interactions through multiple peptide loops that bind to the protein of interest, and the co-operativity between these loops aid in high affinity and selectivity. Those loops are generated after multiple rounds of recombination and somatic hypermutation that helps in identifying the highly selective ligands.
To identify ligands in vitro, targeting a protein of interest, researchers employ either high throughput screening, rational drug design, or display systems such as Phage display or DNA encoded libraries. Each of the methods has its advantages and limitations. Prof. Winssinger’s vision was to generate high-affinity ligands by establishing an in vitro methodology that mimics nature’s antibody selection. This publication introduces a new type of ligand called Suprabody generated using a novel screening methodology. The proposed methodology takes advantage of the current screening technologies and overcomes some of their limitations.
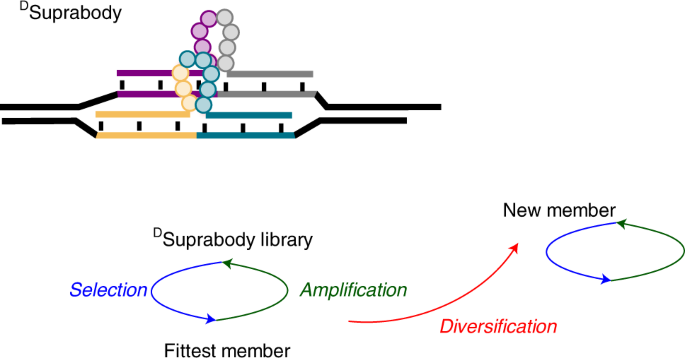
The first step was to generate a Suprabody library of peptide loops coded by Peptide nucleic acids (PNA) at both termini. A PNA-Peptide conjugates library was first synthesized via split and pool technology using solid-phase peptide synthesis. The loops were then generated using the hybridization of the PNA in the PNA-peptide library to a complementary DNA library. Finally, two such DNA-PNA hybrids assemble by the complementarity of their primers to form a DSuprabody library. Thus, DSuprabodies present two constrained peptide loops that can co-operate for binding with the goal of generating high-affinity ligands like antibodies. The use of the DNA template also allows recombination to generate diversity during multiple rounds of selection.
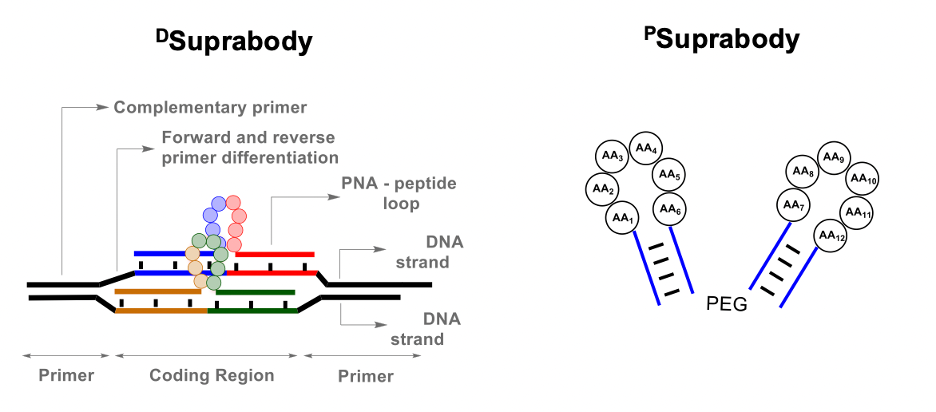
As a proof of principle, we first generated libraries containing the Strep-tag peptide as a control for a selection against Streptavidin. Upon generation of the two PNA and DNA libraries, DSuprabodies assembled well in the tested conditions. The library of DSuprabodies with Strep-tag as one of its members was subjected to screening against immobilized Streptavidin. After multiple rounds of selection, PCR, and sequencing, the analysis revealed the corresponding Strep-tag encoding sequences as the most abundant. In addition, as the rounds progressed, clear enrichment of the dimer Strep-tag DSuprabody over mono-Strep-tag DSuprabody was observed. Finally, the result was further quantified by using qPCR to validate that dimer-Strep-tag DSuprabody was, in fact, better than the mono-Strep-tag DSuprabody. We reconstituted the DSuprabody to a much smaller PSuprabody, generating the constricted peptide loops by using two complementary PNA (5mer or 4mer) sequences and demonstrated that they maintained their binding affinity to Streptavidin.
Having gained confidence in the robustness of the methodology from the model studies, we decided to target a protein-protein interaction with relevance in cancer for what we chose, the Programmed death-ligand1 (PD-L1) protein. Using a library of 108 DSuprabodies, we performed three rounds of selection against an immobilized PD-L1, identifying a PSuprabody that binds to PD-L1 with a binding affinity (Kd) of 70nM. We also stained PD-L1 expressing cells with the selected PSuprabody, and membrane-specific staining was observed.
The results were, in fact, well correlated with mathematical modelling, which shows not only the importance of recombination in generating high-affinity binders but how the false positives and false negatives are minimized, which is one of the critical problems in selections using DEL. While the library used was composed of 108 members, which can be screened by affinity purification, the recombination mechanism allows to screen larger libraries despite not all the library members need to be present in the initial library. In other words, the initial round results in a better binding loop that recombines with other loops in subsequent rounds that eventually aid in providing better binders.
In conclusion, Suprabodies are a new promising class of supramolecules that might be used as drugs, imaging agents, or diagnostic tools. Further investigations are being carried out to develop second-generation Suprabodies where more than two loops are presented, to provide better cooperativity and, therefore, achieve higher affinities.
More Information: https://www.nature.com/articles/s41557-021-00829-5
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in