A Mystery clade in the Tree of Life
Published in Ecology & Evolution
It’s a bird! It’s a plane! No - it’s bacteriophage! This was the line of thinking I had throughout this study. Well sort of - more like thinking from aliens, to a fourth domain of life, then to giant viruses, and, finally, to jumbo bacteriophage. I can explain.
When I first joined the Aylward Lab at Virginia Tech, I was interested in studying viral diversity. My advisor Frank showed me some interesting RNA Polymerase (RNAP) sequences. RNAP is a central component of the transcriptional apparatus, and as such it’s found in all cells - from those of bacteria to those of you and me. RNAP sequences have both enough conservation in all organisms to align them together and enough diversity to distinguish organisms from each other. Thus, these sequences for RNAP are especially useful when constructing a Tree of Life that looks at the diversity of all organisms.
These intriguing RNAP manifested in a Tree of Life that we constructed with RNAP from reference genomes and environmental sequences. In this tree, we saw the usual major branches, or clades, belonging to the three domains of life: Bacteria, Archaea, and Eukarya. Surprisingly, though, this tree had an additional clade that branched away from the other domains. Thus began my mission: to uncover what creatures or entities encoded these intriguing RNAP. Frank suggested they belonged to viruses, but my secret hope was that we discovered aliens or, at least, a fourth domain of life - that was never before seen due to some crazy feature, like existing in another dimension.
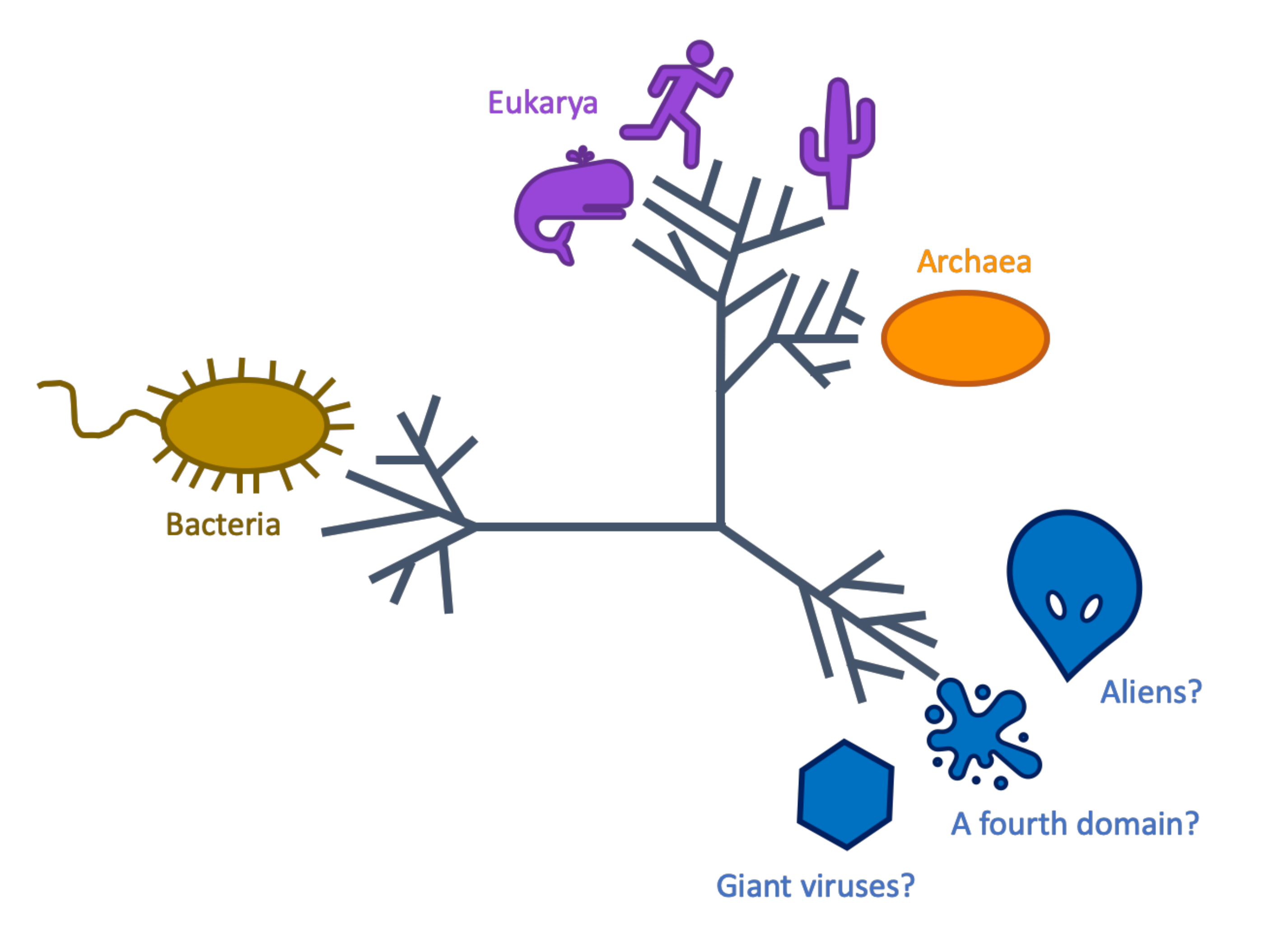
Most known viruses don’t encode their own RNAP because they don’t need it. They can use their host cell’s RNAP during infection. However, a virus having its own RNAP can have advantages, like faster transcription and gene regulation independent of the host. Encoding an RNAP as complex as the one used by cells requires the virus to have room in its genome for such a transcriptional luxury.
Therefore, we suspected these RNAP belonged to the clade of Nucleocytoplasmic Large DNA viruses (NCLDV), in which giant viruses belong. Giant viruses are… giant! They infect eukaryotes and have particle sizes and genome sizes that sometimes exceed those of cells. Many are known to encode their own multi-subunit RNAP. Surprisingly, when I included these giant virus RNAPs in our phylogeny, none of them grouped with that peculiar, ‘alien’ clade. Instead, they grouped near the Eukaryotes and Archaea.
Now nearly convinced that we discovered aliens, my extraterrestrial fantasies were quickly dashed when we considered jumbo bacteriophage, which are viruses of bacteria that have genomes over 200,000 base pairs. Jumbo bacteriophage sometimes encode multi-subunit RNAP, such as the Pseudomonas phage phiKZ. But when I searched cultured bacteriophage genomes for these RNAP, I couldn not even detect them. Their RNAP sequences were too dissimilar from cellular RNAP to align and include in this phylogeny. This result, however, did not mean the mystery clade wasn’t bacteriophage. When we looked at other genes encoded by the mystery clade, we found hallmarks of dsDNA bacteriophage (Caudovirales family), such as tail and terminase proteins. Thus, finally, the case was closed. These RNAP did not belong to a plane, nor a bird, nor giant viruses; they belonged to bacteriophage.
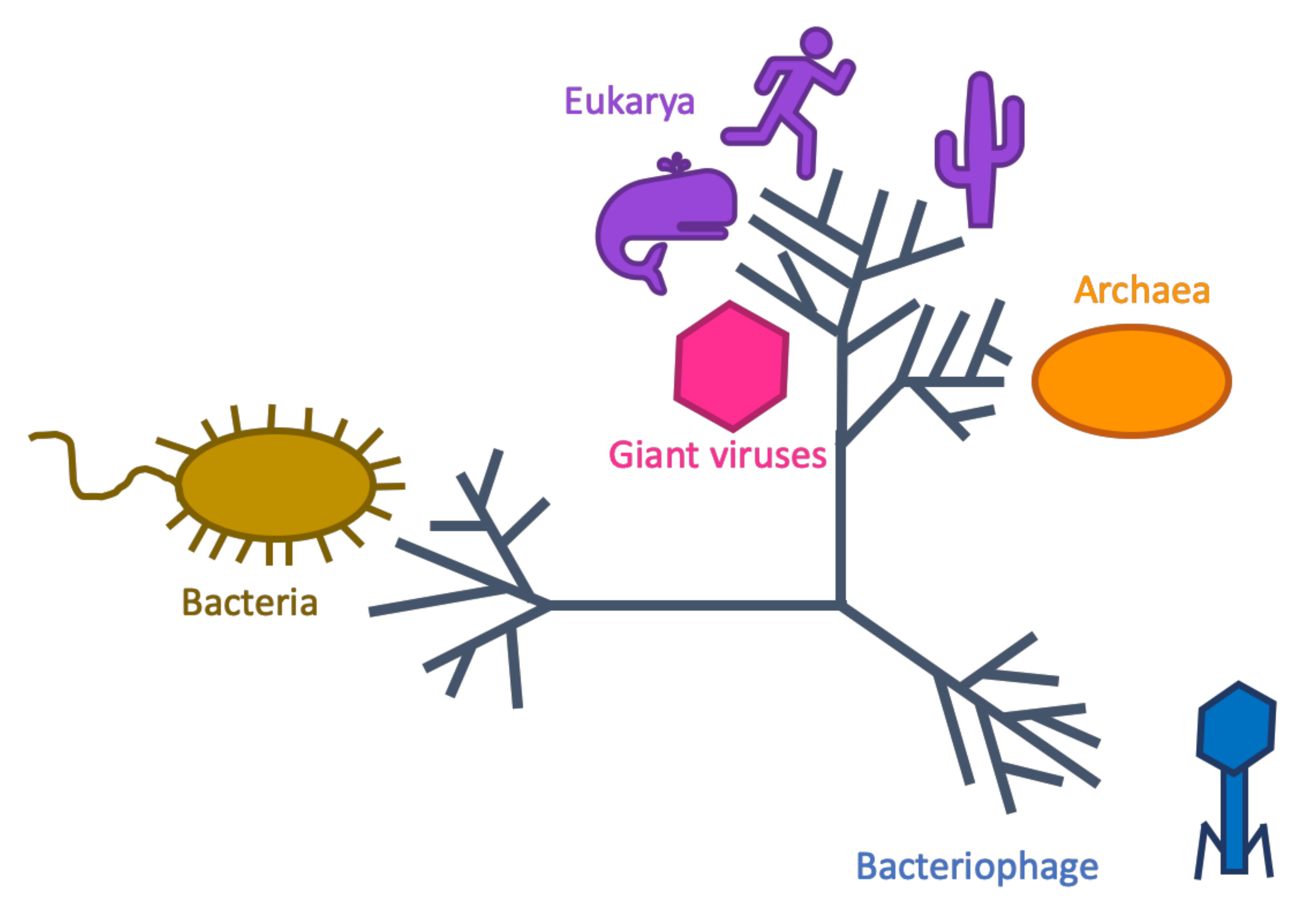
You can read more about these mysterious bacteriophage and the evolutionary implications of their RNAP in our paper: A distinct lineage of Caudovirales that encodes a deeply branching multi-subunit RNA polymerase.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in