Accelerating Diagnoses for Critical Care with Rapid Genome Sequencing
Published in Genetics & Genomics and General & Internal Medicine

Recently, Rapid Whole Genome Sequencing (rWGS) has shown immense promise in molecularly resolving critically ill children. In a recent study involving 184 enrolled infants, 74 (40%) received a diagnosis through rWGS, explaining their admission within a median time of 3 days [1]. In another study, an intensive care unit with both children and adult patients conducted ultrarapid whole genome sequencing and identified diagnostic variants in 5 of the 12 patients (41%) [2]. These patients necessitate rapid progression from sequencing to variant interpretation due to time constraints. Since many of these conditions are intricate and complex, an early genetic disease diagnosis transforms the medical and surgical management of infants in ICUs, ultimately reducing both the duration of empiric interventive treatment and the delay in starting specific treatment.
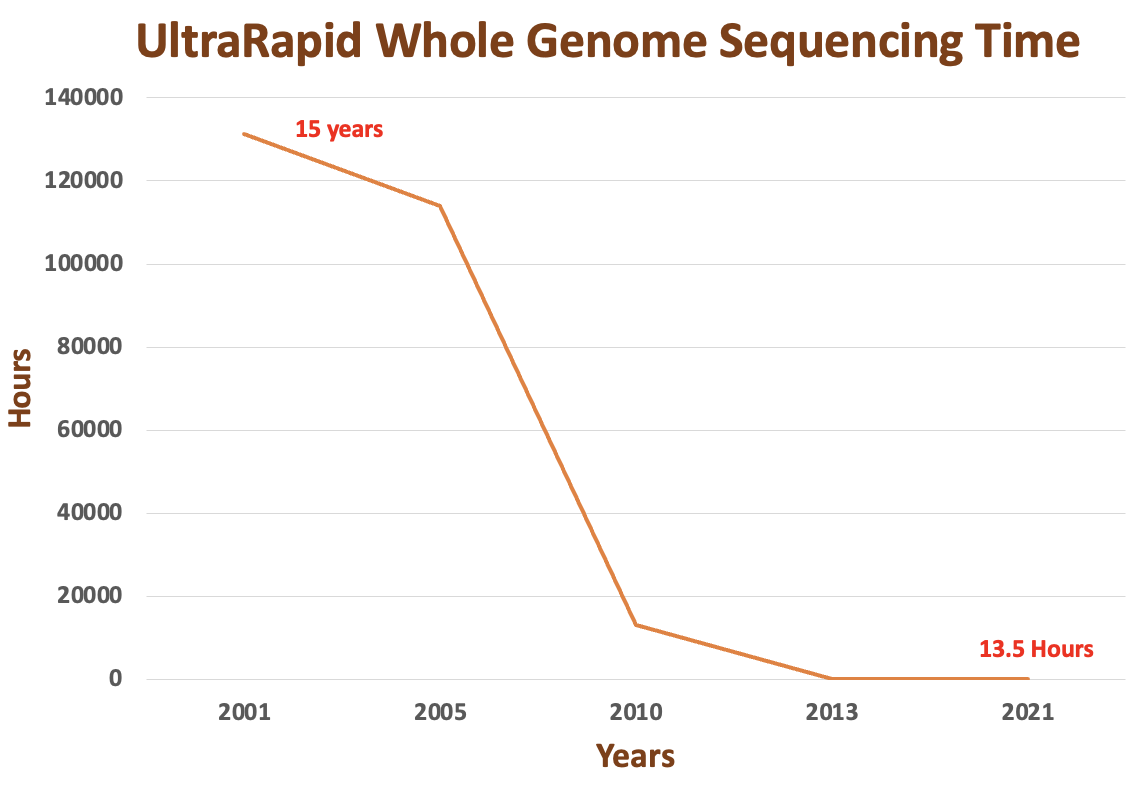
Utilizing various short or long read genome technologies for sequencing can be time-consuming and may not meet the need for speed. Numerous studies have demonstrated the effectiveness of Oxford Nanopore whole genome sequencing, with a turnaround time of within a few hours (less than 10 hours). However, this process involves extensive compute power for base calling, which may not be readily available to regular clinics or hospitals. Conventional sequencing methods may still necessitate a 24-hour timeframe, and an additional day may be required for base calling, mapping, and variant calls. Given the current configuration, it is practically unfeasible for most healthcare institutions to carry out whole genome sequencing within a few hours.
The primary bottleneck lies in variant classification or interpretation based on ACMG guidelines within a reasonable timeframe. Each NICU/PICU case is unique, and predicting the time required to resolve a case with a fast turnaround is challenging. The duration depends on the tools and algorithms employed for variant interpretation. At GenomeArc (https://genomearc.com/), we have developed Horizon, a genomic medicine platform that identifies clinically relevant information from VCF files within 5 minutes for exome and 20 minutes for the whole genome [3]. Horizon provides meaningful diagnostic insights for at least 20-30% of rare disease cohorts. For more complex cases, the Horizon platform generates extensive clinical genomic, pharmacogenomic, and OMICs insights for clinicians and geneticists. In the era of newborn screening and mass population-scale sequencing, ultra-rapid whole genome sequencing is the future, and Horizon is poised to play a significant role in classifying and interpreting genomic variants for diagnosis and therapeutic purposes.
References:




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in