CARPID: Capture RNA-Protein interactions in living cells
Published in Microbiology and Protocols & Methods
Although more than 70% of the genomic DNA can be transcribed to RNA at various stages during development, only ~2% of them codes for protein sequences. For decades, these tremendous number of non-coding RNA species (ncRNA) were recognized as “dark matter” and their function was ignored as transcriptional byproducts or noise. Recent interests have been raised upon these ncRNAs, especially the long non-coding RNAs (lncRNA, defined as ncRNA of more than 200 nucleotides in length) that become widely accepted as important cellular components participating in epigenetic regulation. For example, lncRNA XIST is known to be implicated in X chromosome inactivation in female cells.
For a long period, I have been really curious about the function of these many lncRNAs and the molecular mechanisms of how they are involved in regulating a broad spectrum of biological processes. Identification of the binding proteins of the lncRNA in the cellular context is deemed crucial in unraveling its biological function. Most existing methods rely on the small-molecule or UV-mediated crosslinking, which introduces bias and masks the interacting proteins under physiological circumstances.
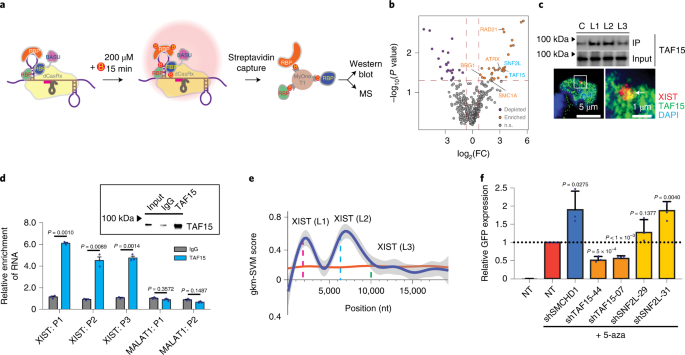
Proximity biotin-labeling technology has been repeatedly used to identify the protein-protein interactions in living cells. A recent publication of a method termed RaPID by the Stanford group led by Dr. Paul A. Khavari using an engineered biotin-transferase BASU to profile the binding proteins of a foreign viral RNA in HEK293T cells inspired me to leverage a similar strategy to identify RNA binding proteins of endogenous lncRNAs of interest. Immediately, I determined to employ the robust RNA-targeting CRISPR/CasRx system to specifically navigate BASU in close proximity to the RNA of interest. We termed the novel method CARPID, short for CRISPR-Assisted RNA-Protein Interaction Detection (See below Figure 1). Powered by the proteomic technique developed by my collaborator Dr Liang Zhang, we were able to find and validate a few previously uncharacterized binding proteins of XIST lncRNA in mammalian cells, e.g. TAF15 and SNF2L, and also demonstrated the broad application of CARPID to lncRNAs in different lengths, abundance and subcellular localizations.
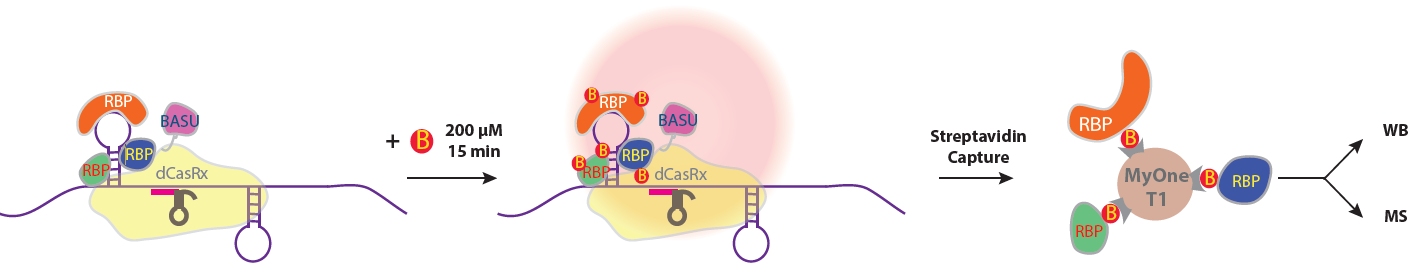
Figure 1 | Scheme of CARPID. The yellow shape depict the navigation system (CRISPR/Cas13d) that uses a 30-nt guide RNA to find and direct the CARPID components to the lncRNA of interest (a purple rope with stem loops). BASU (violate shape attached to CRISPR/Cas13d) is the proximity labeling part of the CARPID that can add biotin (B) moiety to adjacent binding proteins (indicated by “RBP”). Streptavidin coated beads (MyOne T1) is used to recognize and purify the binding proteins (“RBP”) for identification (WB: western blot; MS: mass spectrometry).
During the review process of our manuscript, we also witnessed emergence of a few similar methods from different laboratories. Among them for example, a study led by the Stanford scientist Dr Alice Y. Ting using CasRx-based APEX2 targeting to map RNA-protein interactions was posted in the non-peer reviewed platform bioRxiv, accompanied by another similar publication in Nucleic Acids Research by Zhang et al. from ShanghaiTech University in China using CRISPR/Cas13a to direct a different system PUP-IT for proximity biotin-labeling. All these works strongly encouraged us by showing the broad interest of such strategy. Facing the pandemic of COVID-19, we have been motivated to apply CARPID to the viral RNA genome aiming to identify cellular proteins that bind to the viral RNA and facilitate the life cycle of SARS-CoV-2 in infected cells, which hopefully could enable identification of targets or a path to effectively contain the viral spreading.
CARPID manuscript was first submitted to Nature Methods in September 2019 with two rounds of revision thanks to all very insightful referees, and finally it got accepted on 18th May 2020. During revision, we consecutively faced the social unrest and outbreak of COVID-19 pandemic, which inevitably caused substantial interruption to the lab work. We sincerely wish that the pandemic will be soon controlled and also a peaceful future of Hong Kong and the world.
You can find the online publication from now on via: https://www.nature.com/articles/s41592-020-0866-0
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in